Let-7 expression defines two differentiation stages of cancer
- PMID: 17600087
- PMCID: PMC2040910
- DOI: 10.1073/pnas.0704372104
Let-7 expression defines two differentiation stages of cancer
Abstract
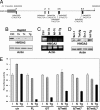
The early phases of carcinogenesis resemble embryonic development, often involving the reexpression of embryonic mesenchymal genes. The NCI60 panel of human tumor cell lines can genetically be subdivided into two superclusters (SCs) that correspond to CD95 Type I and II cells. SC1 cells are characterized by a mesenchymal and SC2 cells by an epithelial gene signature, suggesting that SC1 cells represent less differentiated, advanced stages of cancer. miRNAs are small 20- to 22-nucleotide-long noncoding RNAs that inhibit gene expression at the posttranscriptional level. By performing miRNA expression analysis on 10 Type I and 10 Type II cells, we have determined that SC1 cells express low and SC2 cells high levels of the miRNA let-7, respectively, suggesting that let-7 is a marker for less advanced cancers. Expression of the let-7 target high-mobility group A2 (HMGA2), an early embryonic gene, but not of classical epithelial or mesenchymal markers such as E-cadherin or vimentin, inversely correlated with let-7 expression in SC1 and SC2 cells. Using ovarian cancer as a model, we demonstrate that expression of let-7 and HMGA2 is a better predictor of prognosis than classical markers such as E-cadherin, vimentin, and Snail. These data identify loss of let-7 expression as a marker for less differentiated cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2.Cell Cycle. 2007 Nov 1;6(21):2585-90. doi: 10.4161/cc.6.21.4845. Epub 2007 Aug 6. Cell Cycle. 2007. PMID: 17957144
-
[Construction of let-7d expression vector and its inhibitory effect on HMGA2 and ras expression in human ovarian cancer cells in vitro].Nan Fang Yi Ke Da Xue Xue Bao. 2012 Dec;32(12):1752-7. Nan Fang Yi Ke Da Xue Xue Bao. 2012. PMID: 23268403 Chinese.
-
Regulation of HMGA2 and KRAS genes in epithelial ovarian cancer by miRNA hsa-let-7d-3p.J Cancer Res Ther. 2019 Oct-Dec;15(6):1321-1327. doi: 10.4103/jcrt.JCRT_866_18. J Cancer Res Ther. 2019. PMID: 31898667
-
The role of microRNAs in ovarian cancer.Biomed Res Int. 2014;2014:249393. doi: 10.1155/2014/249393. Epub 2014 Sep 10. Biomed Res Int. 2014. PMID: 25295252 Free PMC article. Review.
-
The role of let-7 in cell differentiation and cancer.Endocr Relat Cancer. 2010 Jan 29;17(1):F19-36. doi: 10.1677/ERC-09-0184. Print 2010 Mar. Endocr Relat Cancer. 2010. PMID: 19779035 Review.
Cited by
-
Molecular deregulation induced by silencing of the high mobility group protein A2 gene in retinoblastoma cells.Mol Vis. 2012;18:2420-37. Epub 2012 Oct 3. Mol Vis. 2012. PMID: 23077401 Free PMC article.
-
Alterations in TGF-β signaling leads to high HMGA2 levels potentially through modulation of PJA1/SMAD3 in HCC cells.Genes Cancer. 2020;11(1-2):43-52. doi: 10.18632/genesandcancer.199. Genes Cancer. 2020. PMID: 32577156 Free PMC article.
-
The Biological Function of MicroRNAs in Bone Tumors.Int J Mol Sci. 2022 Feb 21;23(4):2348. doi: 10.3390/ijms23042348. Int J Mol Sci. 2022. PMID: 35216464 Free PMC article. Review.
-
Combination microRNA-based cellular reprogramming with paclitaxel enhances therapeutic efficacy in a relapsed and multidrug-resistant model of epithelial ovarian cancer.Mol Ther Oncolytics. 2022 Mar 15;25:57-68. doi: 10.1016/j.omto.2022.03.005. eCollection 2022 Jun 16. Mol Ther Oncolytics. 2022. PMID: 35399604 Free PMC article.
-
Identification of target genes for wild type and truncated HMGA2 in mesenchymal stem-like cells.BMC Cancer. 2010 Jun 25;10:329. doi: 10.1186/1471-2407-10-329. BMC Cancer. 2010. PMID: 20576167 Free PMC article.
References
-
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. Cell. 2006;124:1169–1181. - PubMed
-
- Akao Y, Nakagawa Y, Naoe T. Biol Pharm Bull. 2006;29:903–906. - PubMed
-
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Cancer Res. 2004;64:3753–3756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

