Isolation of microRNA targets by miRNP immunopurification
- PMID: 17592038
- PMCID: PMC1924889
- DOI: 10.1261/rna.563707
Isolation of microRNA targets by miRNP immunopurification
Abstract
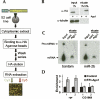
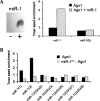
microRNAs (miRNAs) serve as post-transcriptional regulators of gene expression, by guiding effector complexes (miRNPs) to target RNAs. Although considerable progress has been made in computational methods to identify miRNA targets, only a relatively limited assessment of their ability to function in vivo has been reported. Here we describe an alternative approach to miRNA target identification based on a biochemical method for purifying miRNP complexes with associated miRNAs and bound mRNA targets. Microarray analysis revealed a high degree of enrichment for miRNA complementary sites in the 3'UTRs of the miRNP-associated mRNAs. mRNAs specifically associated with an individual miRNA were identified by comparing the miRNP-associated mRNAs from wild-type flies and mutant flies lacking miR-1, and their regulation by the miRNA was validated. This approach provides a means to identify functional miRNA targets based on their physical interaction in vivo.
Figures

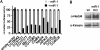
Similar articles
-
Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes.RNA. 2010 Feb;16(2):394-404. doi: 10.1261/rna.1905910. Epub 2009 Dec 30. RNA. 2010. PMID: 20042474 Free PMC article.
-
A miRNA-responsive cell-free translation system facilitates isolation of hepatitis C virus miRNP complexes.RNA. 2013 Aug;19(8):1159-69. doi: 10.1261/rna.038810.113. Epub 2013 Jun 21. RNA. 2013. PMID: 23793894 Free PMC article.
-
Experimental identification of microRNA targets by immunoprecipitation of Argonaute protein complexes.Methods Mol Biol. 2011;732:153-67. doi: 10.1007/978-1-61779-083-6_12. Methods Mol Biol. 2011. PMID: 21431712
-
MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay.Cold Spring Harb Symp Quant Biol. 2006;71:523-30. doi: 10.1101/sqb.2006.71.013. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381335 Review.
-
miRNPs: versatile regulators of gene expression in vertebrate cells.Biochem Soc Trans. 2009 Oct;37(Pt 5):931-5. doi: 10.1042/BST0370931. Biochem Soc Trans. 2009. PMID: 19754429 Review.
Cited by
-
MicroRNA-regulated viral vectors for gene therapy.World J Exp Med. 2016 May 20;6(2):37-54. doi: 10.5493/wjem.v6.i2.37. eCollection 2016 May 20. World J Exp Med. 2016. PMID: 27226955 Free PMC article. Review.
-
Replication competent HIV-1 viruses that express intragenomic microRNA reveal discrete RNA-interference mechanisms that affect viral replication.Cell Biosci. 2011 Nov 23;1(1):38. doi: 10.1186/2045-3701-1-38. Cell Biosci. 2011. PMID: 22112720 Free PMC article.
-
Let-7 microRNAs and Opioid Tolerance.Front Genet. 2012 Jun 21;3:110. doi: 10.3389/fgene.2012.00110. eCollection 2012. Front Genet. 2012. PMID: 22737161 Free PMC article.
-
Identification and characterization of microRNA in the dairy goat (Capra hircus) mammary gland by Solexa deep-sequencing technology.Mol Biol Rep. 2012 Oct;39(10):9361-71. doi: 10.1007/s11033-012-1779-5. Epub 2012 Jul 5. Mol Biol Rep. 2012. PMID: 22763736
-
Comprehensive analysis of microRNA (miRNA) targets in breast cancer cells.J Biol Chem. 2013 Sep 20;288(38):27480-27493. doi: 10.1074/jbc.M113.491803. Epub 2013 Aug 6. J Biol Chem. 2013. PMID: 23921383 Free PMC article.
References
-
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bartel, D.P., Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. - PubMed
-
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell. 2003;113:25–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
