A systematic analysis of the effect of target RNA structure on RNA interference
- PMID: 17576691
- PMCID: PMC1934999
- DOI: 10.1093/nar/gkm437
A systematic analysis of the effect of target RNA structure on RNA interference
Abstract
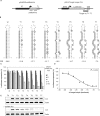
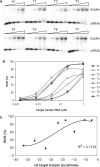
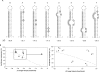
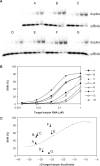
RNAi efficiency is influenced by local RNA structure of the target sequence. We studied this structure-based resistance in detail by targeting a perfect RNA hairpin and subsequently destabilized its tight structure by mutation, thereby gradually exposing the target sequence. Although the tightest RNA hairpins were completely resistant to RNAi, we observed an inverse correlation between the overall target hairpin stability and RNAi efficiency within a specific thermodynamic stability (DeltaG) range. Increased RNAi efficiency was shown to be caused by improved binding of the siRNA to the destabilized target RNA hairpins. The mutational effects vary for different target regions. We find an accessible target 3' end to be most important for RNAi-mediated inhibition. However, these 3' end effects cannot be reproduced in siRNA-target RNA-binding studies in vitro, indicating the important role of RISC components in the in vivo RNAi reaction. The results provide a more detailed insight into the impact of target RNA structure on RNAi and we discuss several possible implications. With respect to lentiviral-mediated delivery of shRNA expression cassettes, we present a DeltaG window to destabilize the shRNA insert for vector improvement, while avoiding RNAi-mediated self-targeting during lentiviral vector production.
Figures
Similar articles
-
HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome.Nucleic Acids Res. 2005 Feb 1;33(2):796-804. doi: 10.1093/nar/gki220. Print 2005. Nucleic Acids Res. 2005. PMID: 15687388 Free PMC article.
-
Local RNA target structure influences siRNA efficacy: a systematic global analysis.J Mol Biol. 2005 May 13;348(4):871-81. doi: 10.1016/j.jmb.2005.03.012. J Mol Biol. 2005. PMID: 15843019
-
Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions.J Mol Biol. 2005 May 13;348(4):883-93. doi: 10.1016/j.jmb.2005.03.011. J Mol Biol. 2005. PMID: 15843020
-
Nonviral vector-mediated RNA interference: its gene silencing characteristics and important factors to achieve RNAi-based gene therapy.Adv Drug Deliv Rev. 2009 Jul 25;61(9):760-6. doi: 10.1016/j.addr.2009.04.006. Epub 2009 Apr 20. Adv Drug Deliv Rev. 2009. PMID: 19386274 Review.
-
siRNA vs. shRNA: similarities and differences.Adv Drug Deliv Rev. 2009 Jul 25;61(9):746-59. doi: 10.1016/j.addr.2009.04.004. Epub 2009 Apr 20. Adv Drug Deliv Rev. 2009. PMID: 19389436 Review.
Cited by
-
AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system.Nucleic Acids Res. 2013 May;41(9):5090-103. doi: 10.1093/nar/gkt193. Epub 2013 Mar 27. Nucleic Acids Res. 2013. PMID: 23535144 Free PMC article.
-
SHAPE-directed discovery of potent shRNA inhibitors of HIV-1.Mol Ther. 2012 Apr;20(4):820-8. doi: 10.1038/mt.2011.299. Epub 2012 Feb 7. Mol Ther. 2012. PMID: 22314289 Free PMC article.
-
Towards Antiviral shRNAs Based on the AgoshRNA Design.PLoS One. 2015 Jun 18;10(6):e0128618. doi: 10.1371/journal.pone.0128618. eCollection 2015. PLoS One. 2015. PMID: 26087209 Free PMC article.
-
RNA Secondary Structure Motifs of the Influenza A Virus as Targets for siRNA-Mediated RNA Interference.Mol Ther Nucleic Acids. 2020 Mar 6;19:627-642. doi: 10.1016/j.omtn.2019.12.018. Epub 2019 Dec 24. Mol Ther Nucleic Acids. 2020. PMID: 31945726 Free PMC article.
-
Ultradeep sequencing analysis of population dynamics of virus escape mutants in RNAi-mediated resistant plants.Mol Biol Evol. 2012 Nov;29(11):3297-307. doi: 10.1093/molbev/mss135. Epub 2012 May 15. Mol Biol Evol. 2012. PMID: 22593223 Free PMC article.
References
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Vance V, Vaucheret H. RNA silencing in plants-defense and counterdefense. Science. 2001;292:2277–2280. - PubMed
-
- Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. - PubMed
-
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 2006;7:590–597. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

