Generation of an inducible and optimized piggyBac transposon system
- PMID: 17576687
- PMCID: PMC1919496
- DOI: 10.1093/nar/gkm446
Generation of an inducible and optimized piggyBac transposon system
Abstract
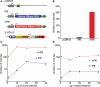
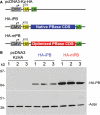
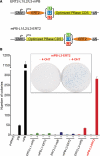
Genomic studies in the mouse have been slowed by the lack of transposon-mediated mutagenesis. However, since the resurrection of Sleeping Beauty (SB), the possibility of performing forward genetics in mice has been reinforced. Recently, piggyBac (PB), a functional transposon from insects, was also described to work in mammals. As the activity of PB is higher than that of SB11 and SB12, two hyperactive SB transposases, we have characterized and improved the PB system in mouse ES cells. We have generated a mouse codon-optimized version of the PB transposase coding sequence (CDS) which provides transposition levels greater than the original. We have also found that the promoter sequence predicted in the 5'-terminal repeat of the PB transposon is active in the mammalian context. Finally, we have engineered inducible versions of the optimized piggyBac transposase fused with ERT2. One of them, when induced, provides higher levels of transposition than the native piggyBac CDS, whereas in the absence of induction its activity is indistinguishable from background. We expect that these tools, adaptable to perform mouse-germline mutagenesis, will facilitate the identification of genes involved in pathological and physiological processes, such as cancer or ES cell differentiation.
Figures

Similar articles
-
Chromosomal transposition of PiggyBac in mouse embryonic stem cells.Proc Natl Acad Sci U S A. 2008 Jul 8;105(27):9290-5. doi: 10.1073/pnas.0801017105. Epub 2008 Jun 25. Proc Natl Acad Sci U S A. 2008. PMID: 18579772 Free PMC article.
-
A hyperactive piggyBac transposase for mammalian applications.Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1531-6. doi: 10.1073/pnas.1008322108. Epub 2011 Jan 4. Proc Natl Acad Sci U S A. 2011. PMID: 21205896 Free PMC article.
-
Hyperactive piggyBac gene transfer in human cells and in vivo.Hum Gene Ther. 2012 Mar;23(3):311-20. doi: 10.1089/hum.2011.138. Epub 2011 Dec 14. Hum Gene Ther. 2012. PMID: 21992617 Free PMC article.
-
Germline mutagenesis mediated by Sleeping Beauty transposon system in mice.Genome Biol. 2007;8 Suppl 1(Suppl 1):S14. doi: 10.1186/gb-2007-8-s1-s14. Genome Biol. 2007. PMID: 18047691 Free PMC article. Review.
-
[The improvement and application of piggyBac transposon system in mammals].Yi Chuan. 2014 Oct;36(10):965-73. doi: 10.3724/SP.J.1005.2014.0965. Yi Chuan. 2014. PMID: 25406244 Review. Chinese.
Cited by
-
A Novel Gene Controls a New Structure: PiggyBac Transposable Element-Derived 1, Unique to Mammals, Controls Mammal-Specific Neuronal Paraspeckles.Mol Biol Evol. 2022 Oct 7;39(10):msac175. doi: 10.1093/molbev/msac175. Mol Biol Evol. 2022. PMID: 36205081 Free PMC article.
-
Potential of transposon-mediated cellular reprogramming towards cell-based therapies.World J Stem Cells. 2020 Jul 26;12(7):527-544. doi: 10.4252/wjsc.v12.i7.527. World J Stem Cells. 2020. PMID: 32843912 Free PMC article. Review.
-
DNA lesion identity drives choice of damage tolerance pathway in murine cell chromosomes.Nucleic Acids Res. 2015 Feb 18;43(3):1637-45. doi: 10.1093/nar/gku1398. Nucleic Acids Res. 2015. PMID: 25589543 Free PMC article.
-
Haploid-genetic screening of trophectoderm specification identifies Dyrk1a as a repressor of totipotent-like status.Sci Adv. 2023 Dec 22;9(51):eadi5683. doi: 10.1126/sciadv.adi5683. Epub 2023 Dec 20. Sci Adv. 2023. PMID: 38117886 Free PMC article.
-
Slingshot: a PiggyBac based transposon system for tamoxifen-inducible 'self-inactivating' insertional mutagenesis.Nucleic Acids Res. 2010 Oct;38(18):e173. doi: 10.1093/nar/gkq658. Epub 2010 Aug 5. Nucleic Acids Res. 2010. PMID: 20688953 Free PMC article.
References
-
- Carlson CM, Largaespada DA. Insertional mutagenesis in mice: new perspectives and tools. Nat. Rev. Genet. 2005;6:568–580. - PubMed
-
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

