An atomic model of the interferon-beta enhanceosome
- PMID: 17574024
- PMCID: PMC2020837
- DOI: 10.1016/j.cell.2007.05.019
An atomic model of the interferon-beta enhanceosome
Abstract
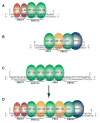
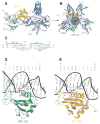
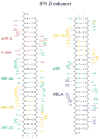
Transcriptional activation of the interferon-beta (IFN-beta) gene requires assembly of an enhanceosome containing ATF-2/c-Jun, IRF-3/IRF-7, and NFkappaB. These factors bind cooperatively to the IFN-beta enhancer and recruit coactivators and chromatin-remodeling proteins to the IFN-beta promoter. We describe here a crystal structure of the DNA-binding domains of IRF-3, IRF-7, and NFkappaB, bound to one half of the enhancer, and use a previously described structure of the remaining half to assemble a complete picture of enhanceosome architecture in the vicinity of the DNA. Association of eight proteins with the enhancer creates a continuous surface for recognizing a composite DNA-binding element. Paucity of local protein-protein contacts suggests that cooperative occupancy of the enhancer comes from both binding-induced changes in DNA conformation and interactions with additional components such as CBP. Contacts with virtually every nucleotide pair account for the evolutionary invariance of the enhancer sequence.
Figures

Similar articles
-
Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer.EMBO J. 2004 Nov 10;23(22):4384-93. doi: 10.1038/sj.emboj.7600453. Epub 2004 Oct 28. EMBO J. 2004. PMID: 15510218 Free PMC article.
-
Assembly of a functional beta interferon enhanceosome is dependent on ATF-2-c-jun heterodimer orientation.Mol Cell Biol. 2000 Jul;20(13):4814-25. doi: 10.1128/MCB.20.13.4814-4825.2000. Mol Cell Biol. 2000. PMID: 10848607 Free PMC article.
-
Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer.Mol Cell. 2007 Jun 8;26(5):703-16. doi: 10.1016/j.molcel.2007.04.022. Mol Cell. 2007. PMID: 17560375
-
The enhanceosome.Curr Opin Struct Biol. 2008 Apr;18(2):236-42. doi: 10.1016/j.sbi.2007.12.002. Epub 2008 Feb 21. Curr Opin Struct Biol. 2008. PMID: 18206362 Review.
-
The transcriptional code of human IFN-beta gene expression.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):328-36. doi: 10.1016/j.bbagrm.2010.01.010. Epub 2010 Jan 30. Biochim Biophys Acta. 2010. PMID: 20116463 Review.
Cited by
-
Human Cytokinome Analysis for Interferon Response.J Virol. 2015 Jul;89(14):7108-19. doi: 10.1128/JVI.03729-14. Epub 2015 Apr 29. J Virol. 2015. PMID: 25926649 Free PMC article.
-
Cross-interference of RLR and TLR signaling pathways modulates antibacterial T cell responses.Nat Immunol. 2012 May 20;13(7):659-66. doi: 10.1038/ni.2307. Nat Immunol. 2012. PMID: 22610141
-
Activation and regulation of interferon-β in immune responses.Immunol Res. 2012 Sep;53(1-3):25-40. doi: 10.1007/s12026-012-8293-7. Immunol Res. 2012. PMID: 22411096 Review.
-
Affinity-optimizing enhancer variants disrupt development.Nature. 2024 Feb;626(7997):151-159. doi: 10.1038/s41586-023-06922-8. Epub 2024 Jan 17. Nature. 2024. PMID: 38233525 Free PMC article.
-
Cell-cell fusion induced by measles virus amplifies the type I interferon response.J Virol. 2007 Dec;81(23):12859-71. doi: 10.1128/JVI.00078-07. Epub 2007 Sep 26. J Virol. 2007. PMID: 17898060 Free PMC article.
References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. - PubMed
-
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. - PubMed
-
- Bannister AJ, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. - PubMed
-
- Berkowitz B, Huang DB, Chen-Park FE, Sigler PB, Ghosh G. The x-ray crystal structure of the NF-kappa B p50. p65 heterodimer bound to the interferon beta -kappa B site. J Biol Chem. 2002;277:24694–24700. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

