Identification of a novel virulence factor in recombinant pneumonia virus of mice
- PMID: 17567693
- PMCID: PMC1951446
- DOI: 10.1128/JVI.00364-07
Identification of a novel virulence factor in recombinant pneumonia virus of mice
Abstract
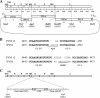
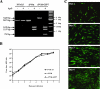
Pneumonia virus of mice (PVM) is a murine relative of human respiratory syncytial virus (HRSV). Here we developed a reverse genetics system for PVM based on a consensus sequence for virulent strain 15. Recombinant PVM and a version engineered to express green fluorescent protein replicated as efficiently as the biological parent in vitro but were 4- and 12.5-fold attenuated in vivo, respectively. The G proteins of HRSV and PVM have been suggested to contribute to viral pathogenesis, but this had not been possible to study in a defined manner in a fully permissive host. As a first step, we evaluated recombinant mutants bearing a deletion of the entire G gene (Delta G) or expressing a G protein lacking its cytoplasmic tail (Gt). Both G mutants replicated as efficiently in vitro as their recombinant parent, but both were nonpathogenic in mice at doses that would otherwise be lethal. We could not detect replication of the Delta G mutant in mice, indicating that its attenuation is based on a severe reduction in the virus load. In contrast, the Gt mutant appeared to replicate as efficiently in mice as its recombinant parent. Thus, the reduction in virulence associated with the Gt mutant could not be accounted for by a reduction in viral replication. These results identified the cytoplasmic tail of G as a virulence factor whose effect is not mediated solely by the viral load. In addition to its intrinsic interest, a recombinant virus that replicates with wild-type-like efficiency but does not cause disease defines optimal properties for vaccine development.
Figures

Similar articles
-
Both nonstructural proteins NS1 and NS2 of pneumonia virus of mice are inhibitors of the interferon type I and type III responses in vivo.J Virol. 2011 May;85(9):4071-84. doi: 10.1128/JVI.01365-10. Epub 2011 Feb 9. J Virol. 2011. PMID: 21307191 Free PMC article.
-
Deletion of nonstructural proteins NS1 and NS2 from pneumonia virus of mice attenuates viral replication and reduces pulmonary cytokine expression and disease.J Virol. 2009 Feb;83(4):1969-80. doi: 10.1128/JVI.02041-08. Epub 2008 Dec 3. J Virol. 2009. PMID: 19052095 Free PMC article.
-
Reevaluation of the virulence of prototypic strain 15 of pneumonia virus of mice.J Virol. 2004 Dec;78(23):13362-5. doi: 10.1128/JVI.78.23.13362-13365.2004. J Virol. 2004. PMID: 15542688 Free PMC article.
-
Recombinant subgroup B human respiratory syncytial virus expressing enhanced green fluorescent protein efficiently replicates in primary human cells and is virulent in cotton rats.J Virol. 2015 Mar;89(5):2849-56. doi: 10.1128/JVI.03587-14. Epub 2014 Dec 24. J Virol. 2015. PMID: 25540371 Free PMC article.
-
The Pneumonia Virus of Mice (PVM) model of acute respiratory infection.Viruses. 2012 Dec;4(12):3494-510. doi: 10.3390/v4123494. Viruses. 2012. PMID: 23342367 Free PMC article. Review.
Cited by
-
Evaluation of pneumonia virus of mice as a possible human pathogen.J Virol. 2012 May;86(10):5829-43. doi: 10.1128/JVI.00163-12. Epub 2012 Mar 21. J Virol. 2012. PMID: 22438539 Free PMC article.
-
Both nonstructural proteins NS1 and NS2 of pneumonia virus of mice are inhibitors of the interferon type I and type III responses in vivo.J Virol. 2011 May;85(9):4071-84. doi: 10.1128/JVI.01365-10. Epub 2011 Feb 9. J Virol. 2011. PMID: 21307191 Free PMC article.
-
Human metapneumovirus glycoprotein G inhibits innate immune responses.PLoS Pathog. 2008 May 30;4(5):e1000077. doi: 10.1371/journal.ppat.1000077. PLoS Pathog. 2008. PMID: 18516301 Free PMC article.
-
S. mansoni bolsters anti-viral immunity in the murine respiratory tract.PLoS One. 2014 Nov 14;9(11):e112469. doi: 10.1371/journal.pone.0112469. eCollection 2014. PLoS One. 2014. PMID: 25398130 Free PMC article.
-
In situ evolution of virus-specific cytotoxic T cell responses in the lung.J Virol. 2013 Oct;87(20):11267-75. doi: 10.1128/JVI.00255-13. Epub 2013 Aug 14. J Virol. 2013. PMID: 23946463 Free PMC article.
References
-
- Biacchesi, S., Q. N. Pham, M. H. Skiadopoulos, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2005. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J. Virol. 79:12608-12613. - PMC - PubMed
-
- Biacchesi, S., M. H. Skiadopoulos, K. C. Tran, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2004. Recovery of human metapneumovirus from cDNA: optimization of growth in vitro and expression of additional genes. Virology 321:247-259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

