Direct measurement of metal ion chelation in the active site of human ferrochelatase
- PMID: 17566985
- PMCID: PMC2396339
- DOI: 10.1021/bi602418e
Direct measurement of metal ion chelation in the active site of human ferrochelatase
Abstract
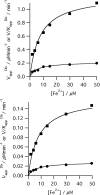
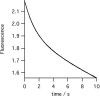
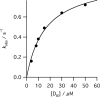
The final step in heme biosynthesis, insertion of ferrous iron into protoporphyrin IX, is catalyzed by protoporphyrin IX ferrochelatase (EC 4.99.1.1). We demonstrate that pre-steady state human ferrochelatase (R115L) exhibits a stoichiometric burst of product formation and substrate consumption, consistent with a rate-determining step following metal ion chelation. Detailed analysis shows that chelation requires at least two steps, rapid binding followed by a slower (k approximately 1 s-1) irreversible step, provisionally assigned to metal ion chelation. Comparison with steady state data reveals that the rate-determining step in the overall reaction, conversion of free porphyrin to free metalloporphyrin, occurs after chelation and is most probably product release. We have measured rate constants for significant steps on the enzyme and demonstrate that metal ion chelation, with a rate constant of 0.96 s-1, is approximately 10 times faster than the rate-determining step in the steady state (kcat = 0.1 s-1). The effect of an additional E343D mutation is apparent at multiple stages in the reaction cycle with a 7-fold decrease in kcat and a 3-fold decrease in kchel. This conservative mutation primarily affects events occurring after metal ion chelation. Further evaluation of structure-function data on site-directed mutants will therefore require both steady state and pre-steady state approaches.
Figures

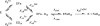

Similar articles
-
Binding of protoporphyrin IX and metal derivatives to the active site of wild-type mouse ferrochelatase at low porphyrin-to-protein ratios.Biochemistry. 2002 Jul 2;41(26):8253-62. doi: 10.1021/bi025569m. Biochemistry. 2002. PMID: 12081474
-
Metal ion selectivity and substrate inhibition in the metal ion chelation catalyzed by human ferrochelatase.J Biol Chem. 2009 Dec 4;284(49):33795-9. doi: 10.1074/jbc.M109.030205. Epub 2009 Sep 19. J Biol Chem. 2009. PMID: 19767646 Free PMC article.
-
Human ferrochelatase: characterization of substrate-iron binding and proton-abstracting residues.Biochemistry. 2001 Aug 21;40(33):9821-7. doi: 10.1021/bi010012c. Biochemistry. 2001. PMID: 11502175
-
Dissection of porphyrin-induced conformational dynamics in the heme biosynthesis enzyme ferrochelatase.Biochemistry. 2012 Sep 11;51(36):7116-27. doi: 10.1021/bi300704c. Epub 2012 Aug 29. Biochemistry. 2012. PMID: 22897320
-
Mechanisms of toxicity of photoactivated artificial porphyrins. Role of porphyrin-protein interactions.Ann N Y Acad Sci. 1987;514:309-22. doi: 10.1111/j.1749-6632.1987.tb48786.x. Ann N Y Acad Sci. 1987. PMID: 3327434 Review. No abstract available.
Cited by
-
Bacterial ferrochelatase turns human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase.J Biol Inorg Chem. 2011 Feb;16(2):235-42. doi: 10.1007/s00775-010-0720-4. Epub 2010 Nov 4. J Biol Inorg Chem. 2011. PMID: 21052751
-
FERROCHELATASE: THE CONVERGENCE OF THE PORPHYRIN BIOSYNTHESIS AND IRON TRANSPORT PATHWAYS.J Porphyr Phthalocyanines. 2011;15(5-6):350-356. doi: 10.1142/S108842461100332X. J Porphyr Phthalocyanines. 2011. PMID: 21852895 Free PMC article.
-
Ferrochelatase: Mapping the Intersection of Iron and Porphyrin Metabolism in the Mitochondria.Front Cell Dev Biol. 2022 May 12;10:894591. doi: 10.3389/fcell.2022.894591. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35646904 Free PMC article. Review.
-
Handling heme: The mechanisms underlying the movement of heme within and between cells.Free Radic Biol Med. 2019 Mar;133:88-100. doi: 10.1016/j.freeradbiomed.2018.08.005. Epub 2018 Aug 6. Free Radic Biol Med. 2019. PMID: 30092350 Free PMC article. Review.
-
The active site of magnesium chelatase.Nat Plants. 2020 Dec;6(12):1491-1502. doi: 10.1038/s41477-020-00806-9. Epub 2020 Nov 30. Nat Plants. 2020. PMID: 33257858
References
-
- Sellers VM, Wu C-K, Dailey TA, Dailey HA. Human ferrochelatase: characterization of substrate-iron binding and proton-abstracting residues. Biochemistry. 2001;40:9821–9827. - PubMed
-
- Dailey HA, Fleming JE. Bovine ferrochelatase. Kinetic analysis of inhibition by N-methylprotoporphyrin, manganese, and heme. Journal of Biological Chemistry. 1983;258:11453–11459. - PubMed
-
- Romesberg FE, Santarsiero BD, Spiller B, Yin J, Barnes D, Schultz PG, Stevens RC. Structural and kinetic evidence for strain in biological catalysis. Biochemistry. 1998;37:14404–14409. - PubMed
-
- Blackwood ME, Jr., Rush TS, III, Medlock A, Dailey HA, Spiro TG. Resonance raman spectra of ferrochelatase reveal porphyrin distortion upon metal binding. Journal of the American Chemical Society. 1997;119:12170–12174.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

