Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein
- PMID: 17547769
- PMCID: PMC1891284
- DOI: 10.1186/1743-422X-4-49
Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein
Abstract
Background: The influenza A virus replicates in the nucleus of its host cell. Thus, entry of the influenza genome into the cell nucleus is necessary for establishing infection. The genome of the influenza A virus consists of eight single-stranded, negative-sense RNA molecules, individually packed with several copies of the viral nucleoprotein (NP) into ribonucleoprotein particles (vRNPs). These vRNPs are large, rod-shaped complexes containing a core of NP, around which the RNA is helically wrapped. The vRNPs are the entities that enter the nucleus, and their nuclear import must be mediated by nuclear localization sequences (NLSs) exposed on the vRNPs. NP contains at least two putative NLSs, one at the N-terminus (NLS1) and one in the middle (NLS2) of the protein. These NP NLSs have been shown to mediate the nuclear import of recombinant NP molecules. However, it remains to be determined which NLS mediates the nuclear import of influenza vRNP complexes.
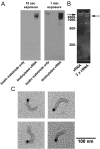
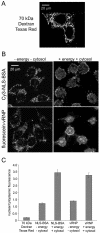
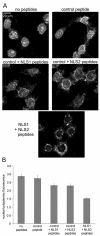
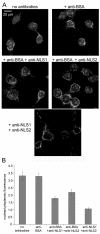
Results: To directly track the nuclear import of the influenza A genome, we developed an experimental assay based on digitonin-permeabilized cells and fluorescently-labeled vRNPs isolated from the influenza A virus. We used this assay to determine the contribution of the two proposed NLSs on NP to the nuclear import of influenza vRNP complexes. Peptides that mimic each of the two NLSs on NP were used to compete with vRNPs for their nuclear import receptors. In addition, antibodies against the two NP NLSs were used to block the NLSs on the vRNP complexes, and thereby inhibit vRNP nuclear import. Both peptide competition and antibody inhibition of either sequence resulted in decreased nuclear accumulation of vRNPs. The two sequences act independently of each other, as inhibition of only one of the two NLSs still resulted in significant, though diminished, nuclear import of vRNPs. Furthermore, when both sequences were blocked, vRNP nuclear import was almost completely inhibited. Antibody inhibition studies further showed that NLS1 on NP is the main contributor to the nuclear import of vRNPs.
Conclusion: Our results demonstrate that both NLS1 and NLS2 on NP can mediate the nuclear uptake of influenza A vRNPs.
Figures

Similar articles
-
The directionality of the nuclear transport of the influenza A genome is driven by selective exposure of nuclear localization sequences on nucleoprotein.Virol J. 2009 Jun 2;6:68. doi: 10.1186/1743-422X-6-68. Virol J. 2009. PMID: 19490630 Free PMC article.
-
Ultrastructural analysis of the nuclear localization sequences on influenza A ribonucleoprotein complexes.J Mol Biol. 2007 Dec 7;374(4):910-6. doi: 10.1016/j.jmb.2007.10.022. Epub 2007 Oct 13. J Mol Biol. 2007. PMID: 17976646
-
An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein.Traffic. 2005 Mar;6(3):205-13. doi: 10.1111/j.1600-0854.2005.00263.x. Traffic. 2005. PMID: 15702989
-
Trafficking of viral genomic RNA into and out of the nucleus: influenza, Thogoto and Borna disease viruses.Virus Res. 2003 Sep;95(1-2):3-12. doi: 10.1016/s0168-1702(03)00159-x. Virus Res. 2003. PMID: 12921991 Review.
-
Biogenesis, assembly, and export of viral messenger ribonucleoproteins in the influenza A virus infected cell.RNA Biol. 2013 Aug;10(8):1274-82. doi: 10.4161/rna.25356. Epub 2013 Jun 17. RNA Biol. 2013. PMID: 23807439 Free PMC article. Review.
Cited by
-
Upregulation of galectin-3 in influenza A virus infection promotes viral RNA synthesis through its association with viral PA protein.J Biomed Sci. 2023 Feb 23;30(1):14. doi: 10.1186/s12929-023-00901-x. J Biomed Sci. 2023. PMID: 36823664 Free PMC article.
-
Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection.Nat Immunol. 2017 Nov;18(11):1249-1260. doi: 10.1038/ni.3837. Epub 2017 Sep 11. Nat Immunol. 2017. PMID: 28892471 Free PMC article.
-
Identification of a putative nuclear localization signal in the tumor suppressor maspin sheds light on its nuclear import regulation.FEBS Open Bio. 2019 Jul;9(7):1174-1183. doi: 10.1002/2211-5463.12626. Epub 2019 May 29. FEBS Open Bio. 2019. PMID: 31144423 Free PMC article.
-
Microtubules in Influenza Virus Entry and Egress.Viruses. 2020 Jan 17;12(1):117. doi: 10.3390/v12010117. Viruses. 2020. PMID: 31963544 Free PMC article. Review.
-
The directionality of the nuclear transport of the influenza A genome is driven by selective exposure of nuclear localization sequences on nucleoprotein.Virol J. 2009 Jun 2;6:68. doi: 10.1186/1743-422X-6-68. Virol J. 2009. PMID: 19490630 Free PMC article.
References
-
- Lamb RA, Krug RM. Orthomyxoviridae: The virus and their replication. In: Knipe, D.M. and Howley PM, editor. Fields Virology. , Lippincott Williams & Wilkins; 2001. pp. 1487–1532.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

