Mamu-B*08-positive macaques control simian immunodeficiency virus replication
- PMID: 17537848
- PMCID: PMC1951344
- DOI: 10.1128/JVI.00895-07
Mamu-B*08-positive macaques control simian immunodeficiency virus replication
Abstract
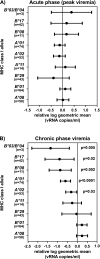
Certain major histocompatibility complex (MHC) class I alleles are associated with the control of human immunodeficiency virus and simian immunodeficiency virus (SIV) replication. We have designed sequence-specific primers for detection of the rhesus macaque MHC class I allele Mamu-B*08 by PCR and screened a cohort of SIV-infected macaques for this allele. Analysis of 196 SIV(mac)239-infected Indian rhesus macaques revealed that Mamu-B*08 was significantly overrepresented in elite controllers; 38% of elite controllers were Mamu-B*08 positive compared to 3% of progressors (P = 0.00001). Mamu-B*08 was also associated with a 7.34-fold decrease in chronic phase viremia (P = 0.002). Mamu-B*08-positive macaques may, therefore, provide a good model to understand the correlates of MHC class I allele-associated immune protection and viral containment in human elite controllers.
Figures

Similar articles
-
Mamu-B*17+ Rhesus Macaques Vaccinated with env, vif, and nef Manifest Early Control of SIVmac239 Replication.J Virol. 2018 Jul 31;92(16):e00690-18. doi: 10.1128/JVI.00690-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875239 Free PMC article.
-
Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication.J Virol. 2008 Feb;82(4):1723-38. doi: 10.1128/JVI.02084-07. Epub 2007 Dec 5. J Virol. 2008. PMID: 18057253 Free PMC article.
-
Escape from CD8(+) T cell responses in Mamu-B*00801(+) macaques differentiates progressors from elite controllers.J Immunol. 2012 Apr 1;188(7):3364-70. doi: 10.4049/jimmunol.1102470. Epub 2012 Mar 2. J Immunol. 2012. PMID: 22387557 Free PMC article.
-
The role of MHC class I gene products in SIV infection of macaques.Immunogenetics. 2017 Aug;69(8-9):511-519. doi: 10.1007/s00251-017-0997-3. Epub 2017 Jul 10. Immunogenetics. 2017. PMID: 28695289 Free PMC article. Review.
-
Making the animal model for AIDS research more precise: the impact of major histocompatibility complex (MHC) genes on pathogenesis and disease progression in SIV-infected monkeys.Curr Mol Med. 2001 Sep;1(4):515-22. doi: 10.2174/1566524013363555. Curr Mol Med. 2001. PMID: 11899095 Review.
Cited by
-
Replicating adenovirus-simian immunodeficiency virus (SIV) recombinant priming and envelope protein boosting elicits localized, mucosal IgA immunity in rhesus macaques correlated with delayed acquisition following a repeated low-dose rectal SIV(mac251) challenge.J Virol. 2012 Apr;86(8):4644-57. doi: 10.1128/JVI.06812-11. Epub 2012 Feb 15. J Virol. 2012. PMID: 22345466 Free PMC article.
-
MHC genotyping from rhesus macaque exome sequences.Immunogenetics. 2019 Sep;71(8-9):531-544. doi: 10.1007/s00251-019-01125-w. Epub 2019 Jul 18. Immunogenetics. 2019. PMID: 31321455 Free PMC article.
-
T-cell correlates of vaccine efficacy after a heterologous simian immunodeficiency virus challenge.J Virol. 2010 May;84(9):4352-65. doi: 10.1128/JVI.02365-09. Epub 2010 Feb 17. J Virol. 2010. PMID: 20164222 Free PMC article.
-
Genomic plasticity of the MHC class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites.Immunogenetics. 2011 Feb;63(2):73-83. doi: 10.1007/s00251-010-0486-4. Epub 2010 Oct 15. Immunogenetics. 2011. PMID: 20949353 Free PMC article.
-
Experimental depletion of CD8+ cells in acutely SIVagm-infected African Green Monkeys results in increased viral replication.Retrovirology. 2010 May 11;7:42. doi: 10.1186/1742-4690-7-42. Retrovirology. 2010. PMID: 20459829 Free PMC article.
References
-
- Allen, T. M., P. Jing, B. Calore, H. Horton, D. H. O'Connor, T. Hanke, M. Piekarczyk, R. Ruddersdorf, B. R. Mothe, C. Emerson, N. Wilson, J. D. Lifson, I. M. Belyakov, J. A. Berzofsky, C. Wang, D. B. Allison, D. C. Montefiori, R. C. Desrosiers, S. Wolinsky, K. J. Kunstman, J. D. Altman, A. Sette, A. J. McMichael, and D. I. Watkins. 2002. Effects of cytotoxic T lymphocytes (CTL) directed against a single simian immunodeficiency virus (SIV) Gag CTL epitope on the course of SIVmac239 infection. J. Virol. 76:10507-10511. - PMC - PubMed
-
- Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 3:e403. - PMC - PubMed
-
- Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H. S. Hewitt, L. Henry, C. H. Linde, J. V. R. Chisholm, T. M. Zaman, E. Pae, S. Mallal, B. D. Walker, A. Sette, B. T. Korber, D. Heckerman, and C. Brander. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094-4101. - PubMed
-
- Bjorkman, P. J., M. A. Saper, B. Samraoui, W. S. Bennett, J. L. Strominger, and D. C. Wiley. 1987. The foreign antigen binding site and T-cell recognition regions of class I histocompatibility antigens. Nature 329:512-518. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- C06 RR020141/RR/NCRR NIH HHS/United States
- C06 RR015459/RR/NCRR NIH HHS/United States
- P51 RR000167/RR/NCRR NIH HHS/United States
- R01 AI052056/AI/NIAID NIH HHS/United States
- R24 RR 016038/RR/NCRR NIH HHS/United States
- R24 RR016038/RR/NCRR NIH HHS/United States
- RR 15459-01/RR/NCRR NIH HHS/United States
- P51 RR 000167/RR/NCRR NIH HHS/United States
- RR 020141-01/RR/NCRR NIH HHS/United States
- R01 AI 049120/AI/NIAID NIH HHS/United States
- Intramural NIH HHS/United States
- R24 RR015371/RR/NCRR NIH HHS/United States
- R01 AI049120/AI/NIAID NIH HHS/United States
- R01 AI 052056/AI/NIAID NIH HHS/United States
- N01CO12400/CA/NCI NIH HHS/United States
- R24 RR 015371/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

