Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity
- PMID: 17533109
- PMCID: PMC2323977
- DOI: 10.1016/j.bbrc.2007.05.092
Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity
Abstract
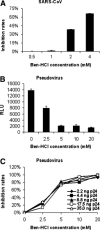
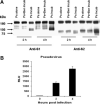
The spike (S) protein of SARS coronavirus (SARS-CoV) has been known to recognize and bind to host receptors, whose conformational changes then facilitate fusion between the viral envelope and host cell membrane, leading to viral entry into target cells. However, other functions of SARS-CoV S protein such as proteolytic cleavage and its implications to viral infection are incompletely understood. In this study, we demonstrated that the infection of SARS-CoV and a pseudovirus bearing the S protein of SARS-CoV was inhibited by a protease inhibitor Ben-HCl. Also, the protease Factor Xa, a target of Ben-HCl abundantly expressed in infected cells, was able to cleave the recombinant and pseudoviral S protein into S1 and S2 subunits, and the cleavage was inhibited by Ben-HCl. Furthermore, this cleavage correlated with the infectivity of the pseudovirus. Taken together, our study suggests a plausible mechanism by which SARS-CoV cleaves its S protein to facilitate viral infection.
Figures

Similar articles
-
The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells.Cell Res. 2004 Oct;14(5):400-6. doi: 10.1038/sj.cr.7290240. Cell Res. 2004. PMID: 15450134 Free PMC article.
-
Roles of spike protein in the pathogenesis of SARS coronavirus.Hong Kong Med J. 2009 Feb;15 Suppl 2:37-40. Hong Kong Med J. 2009. PMID: 19258633
-
[Protease-dependent cell entry mechanism of coronaviruses].Uirusu. 2011 Jun;61(1):109-16. doi: 10.2222/jsv.61.109. Uirusu. 2011. PMID: 21972562 Review. Japanese.
-
Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors.J Virol. 2006 Sep;80(17):8639-52. doi: 10.1128/JVI.00560-06. J Virol. 2006. PMID: 16912312 Free PMC article.
-
The Potential Role of Coagulation Factor Xa in the Pathophysiology of COVID-19: A Role for Anticoagulants as Multimodal Therapeutic Agents.TH Open. 2020 Oct 7;4(4):e288-e299. doi: 10.1055/s-0040-1718415. eCollection 2020 Oct. TH Open. 2020. PMID: 33043235 Free PMC article. Review.
Cited by
-
Coagulopathy in COVID-19: Focus on vascular thrombotic events.J Mol Cell Cardiol. 2020 Sep;146:32-40. doi: 10.1016/j.yjmcc.2020.07.003. Epub 2020 Jul 15. J Mol Cell Cardiol. 2020. PMID: 32681845 Free PMC article. Review.
-
In-stent Thrombosis and COVID-19 Infection: Current Insights on the Mechanistic Relationship.Curr Cardiol Rev. 2023;19(1):e120522204669. doi: 10.2174/1573403X18666220512142019. Curr Cardiol Rev. 2023. PMID: 35549872 Free PMC article. Review.
-
COVID-19-associated diarrhea.World J Gastroenterol. 2021 Jun 21;27(23):3208-3222. doi: 10.3748/wjg.v27.i23.3208. World J Gastroenterol. 2021. PMID: 34163106 Free PMC article. Review.
-
ACE2, COVID-19 Infection, Inflammation, and Coagulopathy: Missing Pieces in the Puzzle.Front Physiol. 2020 Oct 6;11:574753. doi: 10.3389/fphys.2020.574753. eCollection 2020. Front Physiol. 2020. PMID: 33123031 Free PMC article.
-
Host Cell Proteases Involved in Human Respiratory Viral Infections and Their Inhibitors: A Review.Viruses. 2024 Jun 19;16(6):984. doi: 10.3390/v16060984. Viruses. 2024. PMID: 38932275 Free PMC article. Review.
References
-
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. - PMC - PubMed
-
- Mounir S., Talbot P.J. Molecular characterization of the S protein gene of human coronavirus OC43. J. Gen. Virol. 1993;74(Pt. 9):1981–1987. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

