Hinge Atlas: relating protein sequence to sites of structural flexibility
- PMID: 17519025
- PMCID: PMC1913541
- DOI: 10.1186/1471-2105-8-167
Hinge Atlas: relating protein sequence to sites of structural flexibility
Abstract
Background: Relating features of protein sequences to structural hinges is important for identifying domain boundaries, understanding structure-function relationships, and designing flexibility into proteins. Efforts in this field have been hampered by the lack of a proper dataset for studying characteristics of hinges.
Results: Using the Molecular Motions Database we have created a Hinge Atlas of manually annotated hinges and a statistical formalism for calculating the enrichment of various types of residues in these hinges.
Conclusion: We found various correlations between hinges and sequence features. Some of these are expected; for instance, we found that hinges tend to occur on the surface and in coils and turns and to be enriched with small and hydrophilic residues. Others are less obvious and intuitive. In particular, we found that hinges tend to coincide with active sites, but unlike the latter they are not at all conserved in evolution. We evaluate the potential for hinge prediction based on sequence. Motions play an important role in catalysis and protein-ligand interactions. Hinge bending motions comprise the largest class of known motions. Therefore it is important to relate the hinge location to sequence features such as residue type, physicochemical class, secondary structure, solvent exposure, evolutionary conservation, and proximity to active sites. To do this, we first generated the Hinge Atlas, a set of protein motions with the hinge locations manually annotated, and then studied the coincidence of these features with the hinge location. We found that all of the features have bearing on the hinge location. Most interestingly, we found that hinges tend to occur at or near active sites and yet unlike the latter are not conserved. Less surprisingly, we found that hinge residues tend to be small, not hydrophobic or aliphatic, and occur in turns and random coils on the surface. A functional sequence based hinge predictor was made which uses some of the data generated in this study. The Hinge Atlas is made available to the community for further flexibility studies.
Figures
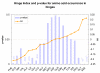
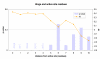
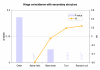
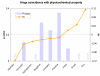
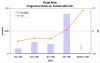

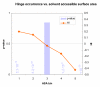
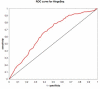

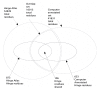
Similar articles
-
FlexOracle: predicting flexible hinges by identification of stable domains.BMC Bioinformatics. 2007 Jun 22;8:215. doi: 10.1186/1471-2105-8-215. BMC Bioinformatics. 2007. PMID: 17587456 Free PMC article.
-
Relating destabilizing regions to known functional sites in proteins.BMC Bioinformatics. 2007 Apr 30;8:141. doi: 10.1186/1471-2105-8-141. BMC Bioinformatics. 2007. PMID: 17470296 Free PMC article.
-
StoneHinge: hinge prediction by network analysis of individual protein structures.Protein Sci. 2009 Feb;18(2):359-71. doi: 10.1002/pro.38. Protein Sci. 2009. PMID: 19180449 Free PMC article.
-
Bioinformatics methods to predict protein structure and function. A practical approach.Mol Biotechnol. 2003 Feb;23(2):139-66. doi: 10.1385/MB:23:2:139. Mol Biotechnol. 2003. PMID: 12632698 Review.
-
Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book.Nat Methods. 2004 Dec;1(3):195-202. doi: 10.1038/nmeth725. Nat Methods. 2004. PMID: 15789030 Review.
Cited by
-
Comparative genome analysis of Mycoplasma pneumoniae.BMC Genomics. 2015 Aug 16;16(1):610. doi: 10.1186/s12864-015-1801-0. BMC Genomics. 2015. PMID: 26275904 Free PMC article.
-
Specificity and Strain-Typing Capabilities of Nanorod Array-Surface Enhanced Raman Spectroscopy for Mycoplasma pneumoniae Detection.PLoS One. 2015 Jun 29;10(6):e0131831. doi: 10.1371/journal.pone.0131831. eCollection 2015. PLoS One. 2015. PMID: 26121242 Free PMC article.
-
Mapping flexibility and the assembly switch of cell division protein FtsZ by computational and mutational approaches.J Biol Chem. 2010 Jul 16;285(29):22554-65. doi: 10.1074/jbc.M110.117127. Epub 2010 May 13. J Biol Chem. 2010. PMID: 20472561 Free PMC article.
-
Sequence analysis of the p1 adhesin gene of Mycoplasma pneumoniae in clinical isolates collected in Beijing in 2008 to 2009.J Clin Microbiol. 2011 Aug;49(8):3000-3. doi: 10.1128/JCM.00105-11. Epub 2011 Jun 22. J Clin Microbiol. 2011. PMID: 21697320 Free PMC article.
-
Conservation of complex knotting and slipknotting patterns in proteins.Proc Natl Acad Sci U S A. 2012 Jun 26;109(26):E1715-23. doi: 10.1073/pnas.1205918109. Epub 2012 Jun 8. Proc Natl Acad Sci U S A. 2012. PMID: 22685208 Free PMC article.
References
-
- The database of macromolecular motions http://www.molmovdb.org
-
- Gerstein RJM, Johnson T, Tsai J, Krebs W. Studying macromolecular motions in a database framework: from structure to sequence. Rigidity Theory and Applications. 1999. pp. 401–442.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

