Intranucleolar sites of ribosome biogenesis defined by the localization of early binding ribosomal proteins
- PMID: 17517959
- PMCID: PMC2064203
- DOI: 10.1083/jcb.200612048
Intranucleolar sites of ribosome biogenesis defined by the localization of early binding ribosomal proteins
Abstract
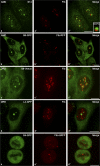
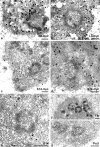
Considerable efforts are being undertaken to elucidate the processes of ribosome biogenesis. Although various preribosomal RNP complexes have been isolated and molecularly characterized, the order of ribosomal protein (r-protein) addition to the emerging ribosome subunits is largely unknown. Furthermore, the correlation between the ribosome assembly pathway and the structural organization of the dedicated ribosome factory, the nucleolus, is not well established. We have analyzed the nucleolar localization of several early binding r-proteins in human cells, applying various methods, including live-cell imaging and electron microscopy. We have located all examined r-proteins (S4, S6, S7, S9, S14, and L4) in the granular component (GC), which is the nucleolar region where later pre-ribosomal RNA (rRNA) processing steps take place. These results imply that early binding r-proteins do not assemble with nascent pre-rRNA transcripts in the dense fibrillar component (DFC), as is generally believed, and provide a link between r-protein assembly and the emergence of distinct granules at the DFC-GC interface.
Figures


Similar articles
-
The functional organization of the nucleolus in proliferating plant cells.Eur J Histochem. 2000;44(2):117-31. Eur J Histochem. 2000. PMID: 10968360 Review.
-
Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP.Mol Cell Biol. 2004 Oct;24(19):8600-10. doi: 10.1128/MCB.24.19.8600-8610.2004. Mol Cell Biol. 2004. PMID: 15367679 Free PMC article.
-
Alterations of nucleolar ultrastructure and ribosome biogenesis by actinomycin D. Implications for U3 snRNP function.Eur J Cell Biol. 1992 Jun;58(1):149-62. Eur J Cell Biol. 1992. PMID: 1386570
-
The location of pKi67 in the outer dense fibrillary compartment of the nucleolus points to a role in ribosome biogenesis during the cell division cycle.J Pathol. 2000 Apr;190(5):537-44. doi: 10.1002/(SICI)1096-9896(200004)190:5<537::AID-PATH577>3.0.CO;2-W. J Pathol. 2000. PMID: 10727979
-
Ribosome biogenesis: of knobs and RNA processing.Exp Cell Res. 2004 May 15;296(1):43-50. doi: 10.1016/j.yexcr.2004.03.016. Exp Cell Res. 2004. PMID: 15120992 Review.
Cited by
-
Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis.J Biol Chem. 2010 Apr 23;285(17):12695-705. doi: 10.1074/jbc.M110.103911. Epub 2010 Feb 16. J Biol Chem. 2010. PMID: 20159986 Free PMC article.
-
Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae.PLoS One. 2013 Jul 10;8(7):e68412. doi: 10.1371/journal.pone.0068412. Print 2013. PLoS One. 2013. PMID: 23874617 Free PMC article.
-
Arabidopsis paralogous genes RPL23aA and RPL23aB encode functionally equivalent proteins.BMC Plant Biol. 2020 Oct 8;20(1):463. doi: 10.1186/s12870-020-02672-1. BMC Plant Biol. 2020. PMID: 33032526 Free PMC article.
-
Aberrant expression of nuclear matrix proteins during HMBA-induced differentiation of gastric cancer cells.World J Gastroenterol. 2010 May 7;16(17):2176-82. doi: 10.3748/wjg.v16.i17.2176. World J Gastroenterol. 2010. PMID: 20440860 Free PMC article.
-
Nucleolus-derived mediators in oncogenic stress response and activation of p53-dependent pathways.Histochem Cell Biol. 2016 Aug;146(2):119-39. doi: 10.1007/s00418-016-1443-6. Epub 2016 May 3. Histochem Cell Biol. 2016. PMID: 27142852 Review.
References
-
- de la Cruz, J., D. Kressler, and P. Linder. 2004. Ribosomal subunit assembly. In The Nucleolus. M. Olson, editor. Landes Bioscience, New York. 258–285.
-
- Decatur, W., and M.J. Fournier. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278:695–698. - PubMed
-
- El Hage, A., and D. Tollervey. 2004. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 1:10–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

