Murine cytomegalovirus major immediate-early enhancer region operating as a genetic switch in bidirectional gene pair transcription
- PMID: 17494084
- PMCID: PMC1933345
- DOI: 10.1128/JVI.02388-06
Murine cytomegalovirus major immediate-early enhancer region operating as a genetic switch in bidirectional gene pair transcription
Abstract
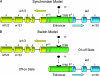
Enhancers are defined as DNA elements that increase transcription when placed in any orientation relative to a promoter. The major immediate-early (MIE) enhancer region of murine cytomegalovirus is flanked by transcription units ie1/3 and ie2, which are transcribed in opposite directions. We have addressed the fundamental mechanistic question of whether the enhancer synchronizes transcription of the bidirectional gene pair (synchronizer model) or whether it operates as a genetic switch, enhancing transcription of either gene in a stochastic alternation (switch model). Clonal analysis of cytokine-triggered, transcription factor-mediated MIE gene expression from latent viral genomes provided evidence in support of the switch model.
Figures


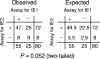
Similar articles
-
Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency.Med Microbiol Immunol. 2008 Jun;197(2):223-31. doi: 10.1007/s00430-007-0069-7. Epub 2007 Dec 19. Med Microbiol Immunol. 2008. PMID: 18097687 Review.
-
Random, asynchronous, and asymmetric transcriptional activity of enhancer-flanking major immediate-early genes ie1/3 and ie2 during murine cytomegalovirus latency in the lungs.J Virol. 2001 Mar;75(6):2692-705. doi: 10.1128/JVI.75.6.2692-2705.2001. J Virol. 2001. PMID: 11222693 Free PMC article.
-
Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice.Virology. 1995 May 10;209(1):236-41. doi: 10.1006/viro.1995.1249. Virology. 1995. PMID: 7747475
-
Rat cytomegalovirus has a unique immediate early gene enhancer.Virology. 1996 Aug 15;222(2):310-7. doi: 10.1006/viro.1996.0428. Virology. 1996. PMID: 8806515
-
To be or not to be active: the stochastic nature of enhancer action.Bioessays. 2000 Apr;22(4):381-7. doi: 10.1002/(SICI)1521-1878(200004)22:4<381::AID-BIES8>3.0.CO;2-E. Bioessays. 2000. PMID: 10723035 Review.
Cited by
-
Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency.Med Microbiol Immunol. 2008 Jun;197(2):223-31. doi: 10.1007/s00430-007-0069-7. Epub 2007 Dec 19. Med Microbiol Immunol. 2008. PMID: 18097687 Review.
-
A temporal gate for viral enhancers to co-opt Toll-like-receptor transcriptional activation pathways upon acute infection.PLoS Pathog. 2015 Apr 9;11(4):e1004737. doi: 10.1371/journal.ppat.1004737. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25856589 Free PMC article.
-
Transactivation of cellular genes involved in nucleotide metabolism by the regulatory IE1 protein of murine cytomegalovirus is not critical for viral replicative fitness in quiescent cells and host tissues.J Virol. 2008 Oct;82(20):9900-16. doi: 10.1128/JVI.00928-08. Epub 2008 Aug 6. J Virol. 2008. PMID: 18684825 Free PMC article.
-
Combinatorial insulin secretion dynamics of recombinant hepatic and enteroendocrine cells.Biotechnol Bioeng. 2012 Apr;109(4):1074-82. doi: 10.1002/bit.24373. Epub 2011 Nov 21. Biotechnol Bioeng. 2012. PMID: 22094821 Free PMC article.
-
The immune evasion paradox: immunoevasins of murine cytomegalovirus enhance priming of CD8 T cells by preventing negative feedback regulation.J Virol. 2008 Dec;82(23):11637-50. doi: 10.1128/JVI.01510-08. Epub 2008 Sep 24. J Virol. 2008. PMID: 18815306 Free PMC article.
References
-
- Adachi, N., and M. R. Lieber. 2002. Bidirectional gene organization: a common architectural feature of the human genome. Cell 109:807-809. - PubMed
-
- Agresti, A. 1992. A survey of exact inference for contingency tables. Stat. Sci. 7:131-177.
-
- Bain, M., M. Reeves, and J. Sinclair. 2006. Regulation of human cytomegalovirus gene expression by chromatin remodeling, p. 167-183. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

