Effect of rapamycin on the fate of P23H opsin associated with retinitis pigmentosa (an American Ophthalmological Society thesis)
- PMID: 17471359
- PMCID: PMC1809918
Effect of rapamycin on the fate of P23H opsin associated with retinitis pigmentosa (an American Ophthalmological Society thesis)
Abstract
Purpose: To determine the effect of rapamycin on the fate of misfolded opsin associated with retinitis pigmentosa.
Methods: Stable cell lines separately expressing WT and P23H opsins and WT and DeltaF508 CFTR were used. Cells were incubated with complete media or amino acid-depleted medium or in the presence of rapamycin. At various time points thereafter, quantitative opsin and CFTR immunoblotting was performed. Immunofluorescence and electron microscopy were also performed to observe the expression and colocalization of autophagy specific marker proteins with opsin or CFTR.
Results: Upon incubation with rapamycin, the levels of P23H opsin and DeltaF508 CFTR were reduced more rapidly than in untreated controls while no observable changes in the amounts of WT opsin was seen. The autophagy specific marker proteins, Atg7, Atg8 (LC3), and LAMP-1, which associate with autophagic vacuoles, colocalized with P23H opsin. A dramatic increase in the immunofluorescence signals of Atg7, LC3, and LAMP-1 was observed. All three of these proteins were found to decorate P23H opsin, suggesting that autophagy may be directly responsible for the clearance of this protein. Also, it was determined that neither the unfolded protein response nor the heat shock response was induced upon rapamycin-associated degradation of P23H opsin.
Conclusions: These data suggest that rapamycin induces the loss of P23H opsin and DeltaF508 CFTR from the cell under the experimental conditions described. Concomitantly, there is increased expression and colocalization of autophagy marker proteins with P23H opsin. Immunogold electron microscopic studies demonstrate autophagic vacuoles clustered in physical proximity to the aggregates of P23H opsin, suggesting that some of the loss of P23H is related to the induction of autophagy. Thus, rapamycin may be useful to clear misfolded proteins associated with retinal degeneration.
Figures



 ) or starved (
) or starved (
 ) or treated with rapamycin (
) or treated with rapamycin (
 ) or starved and treated with rapamycin (
) or starved and treated with rapamycin (
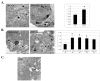
 ). The levels of WT opsin remain essentially unchanged whereas the mutant P23H opsin is lost dramatically. Even the P23H opsin rescued with 11-cis retinal is susceptible to degradation by treatment with rapamycin. Next, HEK 293 cells stably expressing P23H opsin were fed and starved in the presence (
). The levels of WT opsin remain essentially unchanged whereas the mutant P23H opsin is lost dramatically. Even the P23H opsin rescued with 11-cis retinal is susceptible to degradation by treatment with rapamycin. Next, HEK 293 cells stably expressing P23H opsin were fed and starved in the presence (
 ) and absence (
) and absence (
 ) of 3MA, an inhibitor of autophagy (E) and the opsin levels quantified. The upper panel shows the immunoblot of P23H opsin under fed and starved conditions over a time course of 0 to 24 hours. The lower panel shows the immunoblot of P23H opsin after treatment of cells with 3MA in fed and starved medium. Quantification of immunoblot reveals an increase in the relative band intensity of opsin in presence of 3MA. However, P23H opsin-expressing cells when treated with proteasome inhibitor, MG132, under fed and starved conditions for 24 hours (F) showed that MG132 did not significantly reduce the levels of misfolded P23H opsin during starvation. The bar graph shows relative band intensities of P23H opsin under conditions of fed with MG132 (
) of 3MA, an inhibitor of autophagy (E) and the opsin levels quantified. The upper panel shows the immunoblot of P23H opsin under fed and starved conditions over a time course of 0 to 24 hours. The lower panel shows the immunoblot of P23H opsin after treatment of cells with 3MA in fed and starved medium. Quantification of immunoblot reveals an increase in the relative band intensity of opsin in presence of 3MA. However, P23H opsin-expressing cells when treated with proteasome inhibitor, MG132, under fed and starved conditions for 24 hours (F) showed that MG132 did not significantly reduce the levels of misfolded P23H opsin during starvation. The bar graph shows relative band intensities of P23H opsin under conditions of fed with MG132 (
 ) and starved with MG132 (
) and starved with MG132 (
 ).
).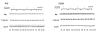

 ) or treated with rapamycin (
) or treated with rapamycin (
 ) or starved and treated with rapamycin (
) or starved and treated with rapamycin (
 ) when compared to fed cells (
) when compared to fed cells (
 ). The levels of WT CFTR remain unchanged over the time course of these conditions.
). The levels of WT CFTR remain unchanged over the time course of these conditions.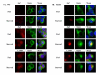
Similar articles
-
Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa.J Biol Chem. 2003 Apr 18;278(16):14442-14450. doi: 10.1074/jbc.M300087200. Epub 2003 Feb 1. J Biol Chem. 2003. PMID: 12566452 Free PMC article.
-
Retinoids assist the cellular folding of the autosomal dominant retinitis pigmentosa opsin mutant P23H.J Biol Chem. 2004 Apr 16;279(16):16278-84. doi: 10.1074/jbc.M312101200. Epub 2004 Feb 9. J Biol Chem. 2004. PMID: 14769795
-
Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa.Hum Mol Genet. 2008 Oct 1;17(19):3043-54. doi: 10.1093/hmg/ddn202. Epub 2008 Jul 17. Hum Mol Genet. 2008. PMID: 18635576
-
Inherent instability of the retinitis pigmentosa P23H mutant opsin.J Biol Chem. 2014 Mar 28;289(13):9288-303. doi: 10.1074/jbc.M114.551713. Epub 2014 Feb 10. J Biol Chem. 2014. PMID: 24515108 Free PMC article.
-
Pharmacological manipulation of rhodopsin retinitis pigmentosa.Adv Exp Med Biol. 2010;664:317-23. doi: 10.1007/978-1-4419-1399-9_36. Adv Exp Med Biol. 2010. PMID: 20238031 Review.
Cited by
-
Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR.Mol Cell. 2014 Apr 10;54(1):166-179. doi: 10.1016/j.molcel.2014.02.025. Epub 2014 Mar 27. Mol Cell. 2014. PMID: 24685158 Free PMC article.
-
The use of induced pluripotent stem cells to reveal pathogenic gene mutations and explore treatments for retinitis pigmentosa.Mol Brain. 2014 Jun 16;7:45. doi: 10.1186/1756-6606-7-45. Mol Brain. 2014. PMID: 24935155 Free PMC article.
-
Autophagy Dysfunction and Oxidative Stress, Two Related Mechanisms Implicated in Retinitis Pigmentosa.Front Physiol. 2018 Jul 26;9:1008. doi: 10.3389/fphys.2018.01008. eCollection 2018. Front Physiol. 2018. PMID: 30093867 Free PMC article. Review.
-
Effects of Epigenetic Modification of PGC-1α by a Chemical Chaperon on Mitochondria Biogenesis and Visual Function in Retinitis Pigmentosa.Cells. 2022 Apr 29;11(9):1497. doi: 10.3390/cells11091497. Cells. 2022. PMID: 35563803 Free PMC article.
-
Autophagy in the eye: implications for ocular cell health.Exp Eye Res. 2014 Jul;124:56-66. doi: 10.1016/j.exer.2014.04.010. Epub 2014 May 6. Exp Eye Res. 2014. PMID: 24810222 Free PMC article. Review.
References
-
- Dryja TP, McGee TL, Reichel E, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. - PubMed
-
- Kaushal S, Khorana HG. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. - PubMed
-
- Illing ME, Rajan RS, Bence NF, Kopito RR. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem. 2002;277:34150–34160. - PubMed
-
- Saliba RS, Munro PM, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci. 2002;115:2907–2918. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
