Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers
- PMID: 17455987
- PMCID: PMC1876407
- DOI: 10.1371/journal.pmed.0040125
Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers
Abstract
Background: The epidermal growth factor receptor (EGFR) gene is the prototype member of the type I receptor tyrosine kinase (TK) family and plays a pivotal role in cell proliferation and differentiation. There are three well described polymorphisms that are associated with increased protein production in experimental systems: a polymorphic dinucleotide repeat (CA simple sequence repeat 1 [CA-SSR1]) in intron one (lower number of repeats) and two single nucleotide polymorphisms (SNPs) in the promoter region, -216 (G/T or T/T) and -191 (C/A or A/A). The objective of this study was to examine distributions of these three polymorphisms and their relationships to each other and to EGFR gene mutations and allelic imbalance (AI) in non-small cell lung cancers.
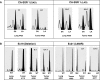
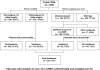
Methods and findings: We examined the frequencies of the three polymorphisms of EGFR in 556 resected lung cancers and corresponding non-malignant lung tissues from 336 East Asians, 213 individuals of Northern European descent, and seven of other ethnicities. We also studied the EGFR gene in 93 corresponding non-malignant lung tissue samples from European-descent patients from Italy and in peripheral blood mononuclear cells from 250 normal healthy US individuals enrolled in epidemiological studies including individuals of European descent, African-Americans, and Mexican-Americans. We sequenced the four exons (18-21) of the TK domain known to harbor activating mutations in tumors and examined the status of the CA-SSR1 alleles (presence of heterozygosity, repeat number of the alleles, and relative amplification of one allele) and allele-specific amplification of mutant tumors as determined by a standardized semiautomated method of microsatellite analysis. Variant forms of SNP -216 (G/T or T/T) and SNP -191 (C/A or A/A) (associated with higher protein production in experimental systems) were less frequent in East Asians than in individuals of other ethnicities (p < 0.001). Both alleles of CA-SSR1 were significantly longer in East Asians than in individuals of other ethnicities (p < 0.001). Expression studies using bronchial epithelial cultures demonstrated a trend towards increased mRNA expression in cultures having the variant SNP -216 G/T or T/T genotypes. Monoallelic amplification of the CA-SSR1 locus was present in 30.6% of the informative cases and occurred more often in individuals of East Asian ethnicity. AI was present in 44.4% (95% confidence interval: 34.1%-54.7%) of mutant tumors compared with 25.9% (20.6%-31.2%) of wild-type tumors (p = 0.002). The shorter allele in tumors with AI in East Asian individuals was selectively amplified (shorter allele dominant) more often in mutant tumors (75.0%, 61.6%-88.4%) than in wild-type tumors (43.5%, 31.8%-55.2%, p = 0.003). In addition, there was a strong positive association between AI ratios of CA-SSR1 alleles and AI of mutant alleles.
Conclusions: The three polymorphisms associated with increased EGFR protein production (shorter CA-SSR1 length and variant forms of SNPs -216 and -191) were found to be rare in East Asians as compared to other ethnicities, suggesting that the cells of East Asians may make relatively less intrinsic EGFR protein. Interestingly, especially in tumors from patients of East Asian ethnicity, EGFR mutations were found to favor the shorter allele of CA-SSR1, and selective amplification of the shorter allele of CA-SSR1 occurred frequently in tumors harboring a mutation. These distinct molecular events targeting the same allele would both be predicted to result in greater EGFR protein production and/or activity. Our findings may help explain to some of the ethnic differences observed in mutational frequencies and responses to TK inhibitors.
Conflict of interest statement
Figures



Similar articles
-
CYP1A1*2A polymorphism as a predictor of clinical outcome in advanced lung cancer patients treated with EGFR-TKI and its combined effects with EGFR intron 1 (CA)n polymorphism.Eur J Cancer. 2011 Sep;47(13):1962-70. doi: 10.1016/j.ejca.2011.04.018. Epub 2011 May 26. Eur J Cancer. 2011. PMID: 21616658
-
Correlations between EGFR gene polymorphisms and pleural metastasis of lung adenocarcinoma.Onco Targets Ther. 2016 Aug 25;9:5257-70. doi: 10.2147/OTT.S97907. eCollection 2016. Onco Targets Ther. 2016. PMID: 27601918 Free PMC article.
-
Shorter EGFR Dinucleotide Repeat Length Predicts Better Response of Patients with Advanced Non-small Cell Lung Cancer to EGFR Tyrosine Kinase Inhibitor.Cell Biochem Biophys. 2015 Dec;73(3):799-804. doi: 10.1007/s12013-014-0289-6. Cell Biochem Biophys. 2015. PMID: 27259328
-
Ethnic differences in response to epidermal growth factor receptor tyrosine kinase inhibitors.J Clin Oncol. 2006 May 10;24(14):2158-63. doi: 10.1200/JCO.2006.06.5961. J Clin Oncol. 2006. PMID: 16682734 Review.
-
East meets West: ethnic differences in epidemiology and clinical behaviors of lung cancer between East Asians and Caucasians.Chin J Cancer. 2011 May;30(5):287-92. doi: 10.5732/cjc.011.10106. Chin J Cancer. 2011. PMID: 21527061 Free PMC article. Review.
Cited by
-
Polymorphisms in intron 1 of the EGFR gene in non-small cell lung cancer patients.Exp Ther Med. 2012 Nov;4(5):785-789. doi: 10.3892/etm.2012.681. Epub 2012 Aug 23. Exp Ther Med. 2012. PMID: 23226726 Free PMC article.
-
Integrating the multiple dimensions of genomic and epigenomic landscapes of cancer.Cancer Metastasis Rev. 2010 Mar;29(1):73-93. doi: 10.1007/s10555-010-9199-2. Cancer Metastasis Rev. 2010. PMID: 20108112 Free PMC article. Review.
-
Pharmacogenomic and pharmacokinetic determinants of erlotinib toxicity.J Clin Oncol. 2008 Mar 1;26(7):1119-27. doi: 10.1200/JCO.2007.13.1128. J Clin Oncol. 2008. PMID: 18309947 Free PMC article.
-
Identifying Asian American lung cancer disparities: A novel analytic approach.JTCVS Open. 2024 Apr 22;20:153-164. doi: 10.1016/j.xjon.2024.04.010. eCollection 2024 Aug. JTCVS Open. 2024. PMID: 39296463 Free PMC article.
-
From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions.Drug Metab Rev. 2008;40(2):187-224. doi: 10.1080/03602530801952864. Drug Metab Rev. 2008. PMID: 18464043 Free PMC article. Review.
References
-
- Arteaga CL, Baselga J. Tyrosine kinase inhibitors: Why does the current process of clinical development not apply to them? Cancer Cell. 2004;5:525–531. - PubMed
-
- Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. - PubMed
-
- Rowinsky EK. The erbB family: Targets for therapeutic development against cancer and therapeutic strategies using monoclonal antibodies and tyrosine kinase inhibitors. Annu Rev Med. 2004;55:433–457. - PubMed
-
- Brattstrom D, Wester K, Bergqvist M, Hesselius P, Malmstrom PU, et al. HER-2, EGFR, COX-2 expression status correlated to microvessel density and survival in resected non-small cell lung cancer. Acta Oncol. 2004;43:80–86. - PubMed
-
- Sozzi G, Miozzo M, Tagliabue E, Calderone C, Lombardi L, et al. Cytogenetic abnormalities and overexpression of receptors for growth factors in normal bronchial epithelium and tumor samples of lung cancer patients. Cancer Res. 1991;51:400–404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

