Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression
- PMID: 17452451
- PMCID: PMC1951505
- DOI: 10.1128/MCB.01789-06
Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression
Abstract
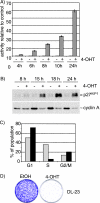
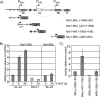
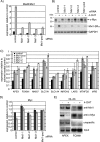
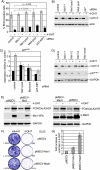
Forkhead transcription factors of the O class (FOXOs) are important targets of the phosphatidylinositol 3-kinase (PI3-kinase)/Akt pathway. FOXOs have been implicated in the regulation of cell cycle progression, oxidative stress resistance, and apoptosis. Using DNA microarrays, we analyzed the transcriptional response to FOXO3a activation by gene expression analysis in DLD-1 colon cancer cells stably expressing a FOXO3a.A3-ER fusion protein. We found that activation of FOXO3a resulted in repression of a number of previously identified Myc target genes. Furthermore, FOXO3a activation induced expression of several members of the Mad/Mxd family of transcriptional repressors, most notably Mxi1. The induction of Mxi1 by FOXO3a was specific to the Mxi1-SR alpha isoform and was mediated by three highly conserved FOXO binding sites within the first intron of the gene. Activation of FOXO3a in response to inhibition of Akt also resulted in activation of Mxi1-SR alpha expression. Silencing of Mxi1 by small interfering RNA (siRNA) reduced FOXO3a-mediated repression of a number of Myc target genes. We also observed that FOXO3a activation induced a switch in promoter occupancy from Myc to Mxi1 on the E-box containing promoter regions of two Myc target genes, APEX and FOXM1. siRNA-mediated transient silencing of Mxi1 or all Mad/Mxd proteins reduced exit from S phase in response to FOXO3a activation, and stable silencing of Mxi1 or Mad1 reduced the growth inhibitory effect of FOXO3a. We conclude that induction of Mad/Mxd proteins contributes to the inhibition of proliferation in response to FOXO3a activation. Our results provide evidence of direct regulation of Mxi1 by FOXO3a and imply an additional mechanism through which the PI3-kinase/Akt/FOXO pathway can modulate Myc function.
Figures




Similar articles
-
c-Myc represses FOXO3a-mediated transcription of the gene encoding the p27(Kip1) cyclin dependent kinase inhibitor.J Cell Biochem. 2008 Aug 15;104(6):2091-106. doi: 10.1002/jcb.21765. J Cell Biochem. 2008. PMID: 18393360
-
Mouse Sin3A interacts with and can functionally substitute for the amino-terminal repression of the Myc antagonist Mxi1.Oncogene. 1996 Mar 7;12(5):1165-72. Oncogene. 1996. PMID: 8649810
-
FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression.Cell Death Differ. 2012 Jun;19(6):968-79. doi: 10.1038/cdd.2011.179. Epub 2011 Dec 2. Cell Death Differ. 2012. PMID: 22139133 Free PMC article.
-
Repression by the Mad(Mxi1)-Sin3 complex.Bioessays. 1998 Oct;20(10):808-18. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. Bioessays. 1998. PMID: 9819568 Review.
-
Two MAD tails: what the recent knockouts of Mad1 and Mxi1 tell us about the MYC/MAX/MAD network.Biochim Biophys Acta. 1999 May 31;1423(3):M37-47. doi: 10.1016/s0304-419x(99)00012-8. Biochim Biophys Acta. 1999. PMID: 10382539 Review.
Cited by
-
Isotretinoin and FoxO1: A scientific hypothesis.Dermatoendocrinol. 2011 Jul;3(3):141-65. doi: 10.4161/derm.3.3.15331. Epub 2011 Jul 1. Dermatoendocrinol. 2011. PMID: 22110774 Free PMC article.
-
FBXO32 Targets c-Myc for Proteasomal Degradation and Inhibits c-Myc Activity.J Biol Chem. 2015 Jun 26;290(26):16202-14. doi: 10.1074/jbc.M115.645978. Epub 2015 May 5. J Biol Chem. 2015. PMID: 25944903 Free PMC article.
-
Linking Metabolic Reprogramming, Plasticity and Tumor Progression.Cancers (Basel). 2021 Feb 12;13(4):762. doi: 10.3390/cancers13040762. Cancers (Basel). 2021. PMID: 33673109 Free PMC article. Review.
-
The "O" class: crafting clinical care with FoxO transcription factors.Adv Exp Med Biol. 2009;665:242-60. doi: 10.1007/978-1-4419-1599-3_18. Adv Exp Med Biol. 2009. PMID: 20429429 Free PMC article. Review.
-
Microbial metabolite restricts 5-fluorouracil-resistant colonic tumor progression by sensitizing drug transporters via regulation of FOXO3-FOXM1 axis.Theranostics. 2022 Jul 18;12(12):5574-5595. doi: 10.7150/thno.70754. eCollection 2022. Theranostics. 2022. PMID: 35910798 Free PMC article.
References
-
- Adhikary, S., and M. Eilers. 2005. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6:635-645. - PubMed
-
- Altomare, D. A., and J. R. Testa. 2005. Perturbations of the AKT signaling pathway in human cancer. Oncogene 24:7455-7464. - PubMed
-
- Bakker, W. J., M. Blazquez-Domingo, A. Kolbus, J. Besooyen, P. Steinlein, H. Beug, P. J. Coffer, B. Lowenberg, M. von Lindern, and T. B. van Dijk. 2004. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J. Cell Biol. 164:175-184. - PMC - PubMed
-
- Barthel, A., D. Schmoll, K. D. Kruger, G. Bahrenberg, R. Walther, R. A. Roth, and H. G. Joost. 2001. Differential regulation of endogenous glucose-6-phosphatase and phosphoenolpyruvate carboxykinase gene expression by the forkhead transcription factor FKHR in H4IIE-hepatoma cells. Biochem. Biophys. Res. Commun. 285:897-902. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
