Portrait of the expansin superfamily in Physcomitrella patens: comparisons with angiosperm expansins
- PMID: 17416912
- PMCID: PMC3243571
- DOI: 10.1093/aob/mcm044
Portrait of the expansin superfamily in Physcomitrella patens: comparisons with angiosperm expansins
Abstract
Background and aims: Expansins are plant cell wall loosening proteins important in a variety of physiological processes. They comprise a large superfamily of genes consisting of four families (EXPA, EXPB, EXLA and EXLB) whose evolutionary relationships have been well characterized in angiosperms, but not in basal land plants. This work attempts to connect the expansin superfamily in bryophytes with the evolutionary history of this superfamily in angiosperms.
Methods: The expansin superfamily in Physcomitrella patens has been assembled from the Physcomitrella sequencing project data generated by the Joint Genome Institute and compared with angiosperm expansin superfamilies. Phylogenetic, motif, intron and distance analyses have been used for this purpose.
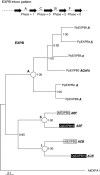
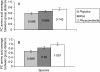
Key results: A gene superfamily is revealed that contains similar numbers of genes as found in arabidopsis, but lacking EXLA or EXLB genes. This similarity in gene numbers exists even though expansin evolution in Physcomitrella diverged from the angiosperm line approx. 400 million years ago. Phylogenetic analyses suggest that there were a minimum of two EXPA genes and one EXPB gene in the last common ancestor of angiosperms and Physcomitrella. Motif analysis seems to suggest that EXPA protein function is similar in bryophytes and angiosperms, but that EXPB function may be altered.
Conclusions: The EXPA genes of Physcomitrella are likely to have maintained the same biochemical function as angiosperm expansins despite their independent evolutionary history. Changes seen at normally conserved residues in the Physcomitrella EXPB family suggest a possible change in function as one mode of evolution in this family.
Figures


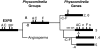


Similar articles
-
Utility of the Amborella trichopoda expansin superfamily in elucidating the history of angiosperm expansins.J Plant Res. 2016 Mar;129(2):199-207. doi: 10.1007/s10265-015-0772-1. Epub 2015 Dec 8. J Plant Res. 2016. PMID: 26646380
-
Selaginella moellendorffii has a reduced and highly conserved expansin superfamily with genes more closely related to angiosperms than to bryophytes.BMC Plant Biol. 2013 Jan 3;13:4. doi: 10.1186/1471-2229-13-4. BMC Plant Biol. 2013. PMID: 23286898 Free PMC article.
-
Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs.J Plant Res. 2006 Jan;119(1):11-21. doi: 10.1007/s10265-005-0253-z. Epub 2006 Jan 13. J Plant Res. 2006. PMID: 16411016 Review.
-
Evolutionary divergence of β-expansin structure and function in grasses parallels emergence of distinctive primary cell wall traits.Plant J. 2015 Jan;81(1):108-20. doi: 10.1111/tpj.12715. Epub 2014 Nov 27. Plant J. 2015. PMID: 25353668
-
Evolutionary crossroads in developmental biology: Physcomitrella patens.Development. 2010 Nov;137(21):3535-43. doi: 10.1242/dev.049023. Development. 2010. PMID: 20940223 Review.
Cited by
-
ABA-Induced Vegetative Diaspore Formation in Physcomitrella patens.Front Plant Sci. 2019 Mar 19;10:315. doi: 10.3389/fpls.2019.00315. eCollection 2019. Front Plant Sci. 2019. PMID: 30941155 Free PMC article.
-
Hydroxyproline O-arabinosyltransferase mutants oppositely alter tip growth in Arabidopsis thaliana and Physcomitrella patens.Plant J. 2016 Jan;85(2):193-208. doi: 10.1111/tpj.13079. Epub 2015 Dec 27. Plant J. 2016. PMID: 26577059 Free PMC article.
-
Novel localization of callose in the spores of Physcomitrella patens and phylogenomics of the callose synthase gene family.Ann Bot. 2009 Mar;103(5):749-56. doi: 10.1093/aob/mcn268. Epub 2009 Jan 19. Ann Bot. 2009. PMID: 19155219 Free PMC article.
-
Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics.PLoS One. 2013 Apr 16;8(4):e62206. doi: 10.1371/journal.pone.0062206. Print 2013. PLoS One. 2013. PMID: 23614035 Free PMC article.
-
Plant expansins: diversity and interactions with plant cell walls.Curr Opin Plant Biol. 2015 Jun;25:162-72. doi: 10.1016/j.pbi.2015.05.014. Epub 2015 Jun 6. Curr Opin Plant Biol. 2015. PMID: 26057089 Free PMC article. Review.

