Protein identification by spectral networks analysis
- PMID: 17404225
- PMCID: PMC1851064
- DOI: 10.1073/pnas.0701130104
Protein identification by spectral networks analysis
Abstract
Advances in tandem mass spectrometry (MS/MS) steadily increase the rate of generation of MS/MS spectra. As a result, the existing approaches that compare spectra against databases are already facing a bottleneck, particularly when interpreting spectra of modified peptides. Here we explore a concept that allows one to perform an MS/MS database search without ever comparing a spectrum against a database. We propose to take advantage of spectral pairs, which are pairs of spectra obtained from overlapping (often nontryptic) peptides or from unmodified and modified versions of the same peptide. Having a spectrum of a modified peptide paired with a spectrum of an unmodified peptide allows one to separate the prefix and suffix ladders, to greatly reduce the number of noise peaks, and to generate a small number of peptide reconstructions that are likely to contain the correct one. The MS/MS database search is thus reduced to extremely fast pattern-matching (rather than time-consuming matching of spectra against databases). In addition to speed, our approach provides a unique paradigm for identifying posttranslational modifications by means of spectral networks analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
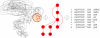
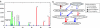

Similar articles
-
An efficient algorithm for the blocked pattern matching problem.Bioinformatics. 2015 Feb 15;31(4):532-8. doi: 10.1093/bioinformatics/btu678. Epub 2014 Oct 15. Bioinformatics. 2015. PMID: 25322837
-
The spectral networks paradigm in high throughput mass spectrometry.Mol Biosyst. 2012 Oct;8(10):2535-44. doi: 10.1039/c2mb25085c. Mol Biosyst. 2012. PMID: 22610447 Free PMC article. Review.
-
Protein Identification from Tandem Mass Spectra by Database Searching.Methods Mol Biol. 2017;1558:357-380. doi: 10.1007/978-1-4939-6783-4_17. Methods Mol Biol. 2017. PMID: 28150247
-
Spectral networks: a new approach to de novo discovery of protein sequences and posttranslational modifications.Biotechniques. 2007 Jun;42(6):687, 689, 691 passim. doi: 10.2144/000112487. Biotechniques. 2007. PMID: 17612289
-
Protein identification by tandem mass spectrometry and sequence database searching.Methods Mol Biol. 2007;367:87-119. doi: 10.1385/1-59745-275-0:87. Methods Mol Biol. 2007. PMID: 17185772 Review.
Cited by
-
Network Topology Evaluation and Transitive Alignments for Molecular Networking.J Am Soc Mass Spectrom. 2024 Sep 4;35(9):2165-2175. doi: 10.1021/jasms.4c00208. Epub 2024 Aug 12. J Am Soc Mass Spectrom. 2024. PMID: 39133821
-
Fast mass spectrometry search and clustering of untargeted metabolomics data.Nat Biotechnol. 2024 Jan 2. doi: 10.1038/s41587-023-01985-4. Online ahead of print. Nat Biotechnol. 2024. PMID: 38168990
-
Fast alignment of mass spectra in large proteomics datasets, capturing dissimilarities arising from multiple complex modifications of peptides.BMC Bioinformatics. 2023 Nov 8;24(1):421. doi: 10.1186/s12859-023-05555-y. BMC Bioinformatics. 2023. PMID: 37940845 Free PMC article.
-
A glimpse into the fungal metabolomic abyss: Novel network analysis reveals relationships between exogenous compounds and their outputs.PNAS Nexus. 2023 Sep 29;2(10):pgad322. doi: 10.1093/pnasnexus/pgad322. eCollection 2023 Oct. PNAS Nexus. 2023. PMID: 37854706 Free PMC article.
-
Combination of GC-MS Molecular Networking and Larvicidal Effect against Aedes aegypti for the Discovery of Bioactive Substances in Commercial Essential Oils.Molecules. 2022 Feb 28;27(5):1588. doi: 10.3390/molecules27051588. Molecules. 2022. PMID: 35268689 Free PMC article.
References
-
- Eng J, McCormack A, Yates J. J Am Soc Mass Spectrom. 1994;5:976–989. - PubMed
-
- Perkins D, Pappin D, Creasy D, Cottrell J. Electrophoresis. 1999;20:3551–3567. - PubMed
-
- Craig R, Beavis R. Bioinformatics. 2004;20:1466–1467. - PubMed
-
- Tanner S, Shu H, Frank A, Wang L, Zandi E, Mumby M, Pevzner P, Bafna V. Anal Chem. 2005;77:4626–4639. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

