Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats
- PMID: 17369305
- PMCID: PMC1913320
- DOI: 10.1128/JB.00187-07
Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats
Abstract
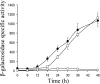
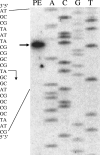
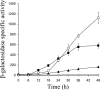
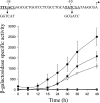
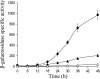
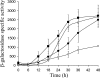
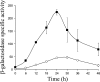
Expression of dev genes is important for triggering spore differentiation inside Myxococcus xanthus fruiting bodies. DNA sequence analysis suggested that dev and cas (CRISPR-associated) genes are cotranscribed at the dev locus, which is adjacent to CRISPR (clustered regularly interspaced short palindromic repeats). Analysis of RNA from developing M. xanthus confirmed that dev and cas genes are cotranscribed with a short upstream gene and at least two repeats of the downstream CRISPR, forming the dev operon. The operon is subject to strong, negative autoregulation during development by DevS. The dev promoter was identified. Its -35 and -10 regions resemble those recognized by M. xanthus sigma(A) RNA polymerase, the homolog of Escherichia coli sigma(70), but the spacer may be too long (20 bp); there is very little expression during growth. Induction during development relies on at least two positive regulatory elements located in the coding region of the next gene upstream. At least two positive regulatory elements and one negative element lie downstream of the dev promoter, such that the region controlling dev expression spans more than 1 kb. The results of testing different fragments for dev promoter activity in wild-type and devS mutant backgrounds strongly suggest that upstream and downstream regulatory elements interact functionally. Strikingly, the 37-bp sequence between the two CRISPR repeats that, minimally, are cotranscribed with dev and cas genes exactly matches a sequence in the bacteriophage Mx8 intP gene, which encodes a form of the integrase needed for lysogenization of M. xanthus.
Figures

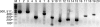
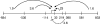

Similar articles
-
The dev Operon Regulates the Timing of Sporulation during Myxococcus xanthus Development.J Bacteriol. 2017 Apr 25;199(10):e00788-16. doi: 10.1128/JB.00788-16. Print 2017 May 15. J Bacteriol. 2017. PMID: 28264995 Free PMC article.
-
devI is an evolutionarily young negative regulator of Myxococcus xanthus development.J Bacteriol. 2015 Apr;197(7):1249-62. doi: 10.1128/JB.02542-14. Epub 2015 Feb 2. J Bacteriol. 2015. PMID: 25645563 Free PMC article.
-
devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus.J Bacteriol. 1993 Nov;175(22):7450-62. doi: 10.1128/jb.175.22.7450-7462.1993. J Bacteriol. 1993. PMID: 7693658 Free PMC article.
-
Highly Signal-Responsive Gene Regulatory Network Governing Myxococcus Development.Trends Genet. 2017 Jan;33(1):3-15. doi: 10.1016/j.tig.2016.10.006. Epub 2016 Dec 2. Trends Genet. 2017. PMID: 27916428 Free PMC article. Review.
-
The complex global response to copper in the multicellular bacterium Myxococcus xanthus.Metallomics. 2018 Jul 18;10(7):876-886. doi: 10.1039/c8mt00121a. Metallomics. 2018. PMID: 29961779 Review.
Cited by
-
Identification of Functions Affecting Predator-Prey Interactions between Myxococcus xanthus and Bacillus subtilis.J Bacteriol. 2016 Nov 18;198(24):3335-3344. doi: 10.1128/JB.00575-16. Print 2016 Dec 15. J Bacteriol. 2016. PMID: 27698086 Free PMC article.
-
Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus.J Bacteriol. 2008 Feb;190(4):1401-12. doi: 10.1128/JB.01415-07. Epub 2007 Dec 7. J Bacteriol. 2008. PMID: 18065539 Free PMC article.
-
Occurrence and Diversity of CRISPR Loci in Lactobacillus casei Group.Front Microbiol. 2020 Apr 8;11:624. doi: 10.3389/fmicb.2020.00624. eCollection 2020. Front Microbiol. 2020. PMID: 32322250 Free PMC article.
-
Two-Component Signal Transduction Systems That Regulate the Temporal and Spatial Expression of Myxococcus xanthus Sporulation Genes.J Bacteriol. 2015 Sep 14;198(3):377-85. doi: 10.1128/JB.00474-15. Print 2016 Feb 1. J Bacteriol. 2015. PMID: 26369581 Free PMC article. Review.
-
Identification of RNA Binding Partners of CRISPR-Cas Proteins in Prokaryotes Using RIP-Seq.Methods Mol Biol. 2022;2404:111-133. doi: 10.1007/978-1-0716-1851-6_6. Methods Mol Biol. 2022. PMID: 34694606
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Ansari, A. Z., J. E. Bradner, and T. V. O'Halloran. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371-375. - PubMed
-
- Arnosti, D. N., and M. M. Kulkarni. 2005. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94:890-898. - PubMed
-
- Biran, D., and L. Kroos. 1997. In vitro transcription of Myxococcus xanthus genes with RNA polymerase containing σA, the major sigma factor in growing cells. Mol. Microbiol. 25:463-472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

