Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production
- PMID: 17360756
- PMCID: PMC1900231
- DOI: 10.1128/JVI.01511-06
Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production
Abstract
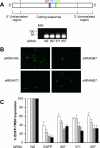
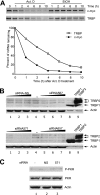
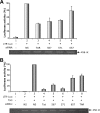
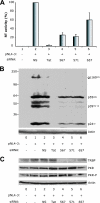
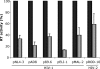
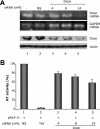
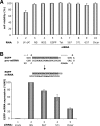
RNA interference (RNAi) is now widely used for gene silencing in mammalian cells. The mechanism uses the RNA-induced silencing complex, in which Dicer, Ago2, and the human immunodeficiency virus type 1 (HIV-1) TAR RNA binding protein (TRBP) are the main components. TRBP is a protein that increases HIV-1 expression and replication by inhibition of the interferon-induced protein kinase PKR and by increasing translation of viral mRNA. After HIV infection, TRBP could restrict the viral RNA through its activity in RNAi or could contribute more to the enhancement of viral replication. To determine which function will be predominant in the virological context, we analyzed whether the inhibition of its expression could enhance or decrease HIV replication. We have generated small interfering RNAs (siRNAs) against TRBP and found that they decrease HIV-1 long terminal repeat (LTR) basal expression 2-fold, and the LTR Tat transactivated level up to 10-fold. In the context of HIV replication, siRNAs against TRBP decrease the expression of viral genes and inhibit viral production up to fivefold. The moderate increase in PKR expression and activation indicates that it contributes partially to viral gene inhibition. The moderate decrease in micro-RNA (miRNA) biogenesis by TRBP siRNAs suggests that in the context of HIV replication, TRBP functions other than RNAi are predominant. In addition, siRNAs against Dicer decrease viral production twofold and impede miRNA biogenesis. These results suggest that, in the context of HIV replication, TRBP contributes mainly to the enhancement of virus production and that Dicer does not mediate HIV restriction by RNAi.
Figures
Similar articles
-
Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity?Retrovirology. 2005 Oct 27;2:65. doi: 10.1186/1742-4690-2-65. Retrovirology. 2005. PMID: 16253139 Free PMC article.
-
HIV-1 RRE RNA acts as an RNA silencing suppressor by competing with TRBP-bound siRNAs.RNA Biol. 2015;12(2):123-35. doi: 10.1080/15476286.2015.1014759. RNA Biol. 2015. PMID: 25668122 Free PMC article.
-
TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing.EMBO Rep. 2005 Oct;6(10):961-7. doi: 10.1038/sj.embor.7400509. EMBO Rep. 2005. PMID: 16142218 Free PMC article.
-
Control of HIV-1 replication by RNA interference.Virus Res. 2004 Jun 1;102(1):53-8. doi: 10.1016/j.virusres.2004.01.015. Virus Res. 2004. PMID: 15068880 Review.
-
[RNA interference and its application in inhibiting HIV-1 infection].Sheng Wu Gong Cheng Xue Bao. 2005 Jul;21(4):516-9. Sheng Wu Gong Cheng Xue Bao. 2005. PMID: 16176084 Review. Chinese.
Cited by
-
The A-rich RNA sequences of HIV-1 pol are important for the synthesis of viral cDNA.Nucleic Acids Res. 2009 Feb;37(3):945-56. doi: 10.1093/nar/gkn1015. Epub 2008 Dec 23. Nucleic Acids Res. 2009. PMID: 19106143 Free PMC article.
-
MicroRNAs and their potential involvement in HIV infection.Trends Pharmacol Sci. 2011 Nov;32(11):675-81. doi: 10.1016/j.tips.2011.07.003. Epub 2011 Aug 19. Trends Pharmacol Sci. 2011. PMID: 21862142 Free PMC article. Review.
-
Shedding Light on the Role of Extracellular Vesicles in HIV Infection and Wound Healing.Viruses. 2020 May 27;12(6):584. doi: 10.3390/v12060584. Viruses. 2020. PMID: 32471020 Free PMC article. Review.
-
Expression of TARBP1 protein in human non-small-cell lung cancer and its prognostic significance.Oncol Lett. 2018 May;15(5):7182-7190. doi: 10.3892/ol.2018.8202. Epub 2018 Mar 7. Oncol Lett. 2018. PMID: 29731880 Free PMC article.
-
Expression profiling of human milk derived exosomal microRNAs and their targets in HIV-1 infected mothers.Sci Rep. 2020 Jul 31;10(1):12931. doi: 10.1038/s41598-020-69799-x. Sci Rep. 2020. PMID: 32737406 Free PMC article. Clinical Trial.
References
-
- Bannwarth, S., and A. Gatignol. 2005. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr. HIV Res. 3:61-71. - PubMed
-
- Bannwarth, S., S. Lainé, A. Daher, N. Grandvaux, G. Clerzius, A. C. LeBlanc, J. Hiscott, and A. Gatignol. 2006. Cell-specific regulation of TRBP1 promoter by NF-Y transcription factor in lymphocytes and astrocytes. J. Mol. Biol. 355:898-910. - PubMed
-
- Bannwarth, S., L. Talakoub, F. Letourneur, M. Duarte, D. F. Purcell, J. Hiscott, and A. Gatignol. 2001. Organization of the human tarbp2 gene reveals two promoters that are repressed in an astrocytic cell line. J. Biol. Chem. 276:48803-48813. - PubMed
-
- Bass, B. L. 2001. RNA interference. The short answer. Nature 411:428-429. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

