Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response
- PMID: 17360504
- PMCID: PMC1820736
- DOI: 10.1073/pnas.0609357104
Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response
Erratum in
- Proc Natl Acad Sci U S A. 2007 Apr 24;104(17):7307
Abstract
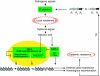
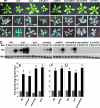
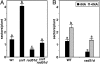
The plant immune response known as systemic acquired resistance (SAR) is a general defense mechanism that confers long-lasting resistance against a broad spectrum of pathogens. SAR triggers many molecular changes including accumulation of antimicrobial pathogenesis-related (PR) proteins. Transcription of PR genes in Arabidopsis is regulated by the coactivator NPR1 and the repressor SNI1. Pathogen infection also triggers an increase in somatic DNA recombination, which results in transmission of changes to the offspring of infected plants. However, it is not known how the induction of homologous recombination during SAR is controlled. Here, we show that SNI1 and RAD51D regulate both gene expression and DNA recombination. In a genetic screen for suppressors of sni1, we discovered that RAD51D is required for NPR1-independent PR gene expression. As a result, the rad51d mutant has enhanced disease susceptibility. Besides altered PR gene expression, rad51d plants are hypersensitive to DNA-damaging agents and are impaired in homologous recombination. The dual role of RAD51D and SNI1 in PR gene transcription and DNA recombination suggests a mechanistic link between the short-term defense response and a long-term survival strategy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

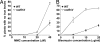
Similar articles
-
DNA repair proteins are directly involved in regulation of gene expression during plant immune response.Cell Host Microbe. 2011 Feb 17;9(2):115-24. doi: 10.1016/j.chom.2011.01.011. Cell Host Microbe. 2011. PMID: 21320694
-
A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity.Plant Cell. 2006 Jul;18(7):1750-65. doi: 10.1105/tpc.105.039677. Epub 2006 Jun 9. Plant Cell. 2006. PMID: 16766691 Free PMC article.
-
Influence of Human p53 on Plant Development.PLoS One. 2016 Sep 20;11(9):e0162840. doi: 10.1371/journal.pone.0162840. eCollection 2016. PLoS One. 2016. PMID: 27648563 Free PMC article.
-
NPR1: the spider in the web of induced resistance signaling pathways.Curr Opin Plant Biol. 2004 Aug;7(4):456-64. doi: 10.1016/j.pbi.2004.05.006. Curr Opin Plant Biol. 2004. PMID: 15231270 Review.
-
[Nonexpressor of pathogenesis-related genes 1 (NPR1): a key node of plant disease resistance signalling network].Sheng Wu Gong Cheng Xue Bao. 2005 Jul;21(4):511-5. Sheng Wu Gong Cheng Xue Bao. 2005. PMID: 16176083 Review. Chinese.
Cited by
-
Plant hormone signaling and modulation of DNA repair under stressful conditions.Plant Cell Rep. 2013 Jul;32(7):1043-52. doi: 10.1007/s00299-013-1410-9. Epub 2013 Mar 19. Plant Cell Rep. 2013. PMID: 23508254 Review.
-
The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells.J Exp Bot. 2012 Sep;63(14):5323-35. doi: 10.1093/jxb/ers190. Epub 2012 Aug 1. J Exp Bot. 2012. PMID: 22859673 Free PMC article.
-
The tRNA thiolation-mediated translational control is essential for plant immunity.Elife. 2024 Jan 29;13:e93517. doi: 10.7554/eLife.93517. Elife. 2024. PMID: 38284752 Free PMC article.
-
A NAC transcription factor and SNI1 cooperatively suppress basal pathogen resistance in Arabidopsis thaliana.Nucleic Acids Res. 2012 Oct;40(18):9182-92. doi: 10.1093/nar/gks683. Epub 2012 Jul 22. Nucleic Acids Res. 2012. PMID: 22826500 Free PMC article.
-
Perspectives of genomic diversification and molecular recombination towards R-gene evolution in plants.Physiol Mol Biol Plants. 2013 Jan;19(1):1-9. doi: 10.1007/s12298-012-0138-2. Physiol Mol Biol Plants. 2013. PMID: 24381433 Free PMC article. Review.
References
-
- Durrant WE, Dong X. Annu Rev Phytopathol. 2004;42:185–209. - PubMed
-
- Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B. Nature. 2003;423:760–762. - PubMed
-
- Molinier J, Ries G, Zipfel C, Hohn B. Nature. 2006;442:1046–1049. - PubMed
-
- Schuermann D, Molinier J, Fritsch O, Hohn B. Trends Genet. 2005;21:172–181. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

