MafB is required for islet beta cell maturation
- PMID: 17360442
- PMCID: PMC1803762
- DOI: 10.1073/pnas.0700013104
MafB is required for islet beta cell maturation
Abstract
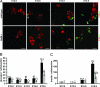
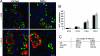
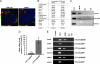
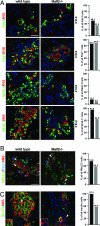
Pancreatic endocrine cell differentiation depends on transcription factors that also contribute in adult insulin and glucagon gene expression. Islet cell development was examined in mice lacking MafB, a transcription factor expressed in immature alpha (glucagon(+)) and beta (insulin(+)) cells and capable of activating insulin and glucagon expression in vitro. We observed that MafB(-/-) embryos had reduced numbers of insulin(+) and glucagon(+) cells throughout development, whereas the total number of endocrine cells was unchanged. Moreover, production of insulin(+) cells was delayed until embryonic day (E) 13.5 in mutant mice and coincided with the onset of MafA expression, a MafB-related activator of insulin transcription. MafA expression was only detected in the insulin(+) cell population in MafB mutants, whereas many important regulatory proteins continued to be expressed in insulin(-) beta cells. However, Pdx1, Nkx6.1, and GLUT2 were selectively lost in these insulin-deficient cells between E15.5 and E18.5. MafB appears to directly regulate transcription of these genes, because binding was observed within endogenous control region sequences. These results demonstrate that MafB plays a previously uncharacterized role by regulating transcription of key factors during development that are required for the production of mature alpha and beta cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Preferential reduction of beta cells derived from Pax6-MafB pathway in MafB deficient mice.Dev Biol. 2008 Feb 15;314(2):443-56. doi: 10.1016/j.ydbio.2007.12.009. Epub 2007 Dec 23. Dev Biol. 2008. PMID: 18199433 Free PMC article.
-
A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells.Dev Biol. 2006 May 15;293(2):526-39. doi: 10.1016/j.ydbio.2006.02.028. Epub 2006 Apr 3. Dev Biol. 2006. PMID: 16580660 Free PMC article.
-
Role of large MAF transcription factors in the mouse endocrine pancreas.Exp Anim. 2015;64(3):305-12. doi: 10.1538/expanim.15-0001. Epub 2015 Apr 27. Exp Anim. 2015. PMID: 25912440 Free PMC article.
-
MafA and MafB activity in pancreatic β cells.Trends Endocrinol Metab. 2011 Sep;22(9):364-73. doi: 10.1016/j.tem.2011.05.003. Epub 2011 Jun 28. Trends Endocrinol Metab. 2011. PMID: 21719305 Free PMC article. Review.
-
Crucial role of PDX-1 in pancreas development, beta-cell differentiation, and induction of surrogate beta-cells.Curr Med Chem. 2007;14(16):1745-52. doi: 10.2174/092986707781058887. Curr Med Chem. 2007. PMID: 17627512 Review.
Cited by
-
Reprogramming adult human dermal fibroblasts to islet-like cells by epigenetic modification coupled to transcription factor modulation.Stem Cells Dev. 2013 Sep 15;22(18):2551-60. doi: 10.1089/scd.2013.0134. Epub 2013 Jun 4. Stem Cells Dev. 2013. PMID: 23627894 Free PMC article.
-
Age-Dependent Pancreatic Gene Regulation Reveals Mechanisms Governing Human β Cell Function.Cell Metab. 2016 May 10;23(5):909-20. doi: 10.1016/j.cmet.2016.04.002. Epub 2016 Apr 28. Cell Metab. 2016. PMID: 27133132 Free PMC article.
-
A novel molecular mechanism involved in multiple myeloma development revealed by targeting MafB to haematopoietic progenitors.EMBO J. 2012 Sep 12;31(18):3704-17. doi: 10.1038/emboj.2012.227. Epub 2012 Aug 17. EMBO J. 2012. PMID: 22903061 Free PMC article.
-
Pancreas cell fate.Birth Defects Res C Embryo Today. 2009 Sep;87(3):232-48. doi: 10.1002/bdrc.20156. Birth Defects Res C Embryo Today. 2009. PMID: 19750517 Free PMC article. Review.
-
Pax6 controls the expression of critical genes involved in pancreatic {alpha} cell differentiation and function.J Biol Chem. 2010 Oct 22;285(43):33381-33393. doi: 10.1074/jbc.M110.147215. Epub 2010 Jun 30. J Biol Chem. 2010. PMID: 20592023 Free PMC article.
References
-
- Steiner DF, Bell GI, Rubenstein AH, Chan SJ. In: Endocrinology. DeGroot LJ, Jameson JL, editors. Philadelphia: Saunders; 2001. pp. 667–696.
-
- Jonsson J, Carlsson L, Edlund T, Edlund H. Nature. 1994;371:606–609. - PubMed
-
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. Development (Cambridge, UK) 1996;122:983–995. - PubMed
-
- Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genes Dev. 1997;11:1662–1673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

