Towards a comprehensive structural coverage of completed genomes: a structural genomics viewpoint
- PMID: 17349043
- PMCID: PMC1829165
- DOI: 10.1186/1471-2105-8-86
Towards a comprehensive structural coverage of completed genomes: a structural genomics viewpoint
Abstract
Background: Structural genomics initiatives were established with the aim of solving protein structures on a large-scale. For many initiatives, such as the Protein Structure Initiative (PSI), the primary aim of target selection is focussed towards structurally characterising protein families which, so far, lack a structural representative. It is therefore of considerable interest to gain insights into the number and distribution of these families, and what efforts may be required to achieve a comprehensive structural coverage across all protein families.
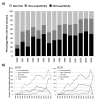
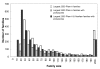
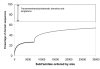
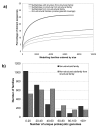
Results: In this analysis we have derived a comprehensive domain annotation of the genomes using CATH, Pfam-A and Newfam domain families. We consider what proportions of structurally uncharacterized families are accessible to high-throughput structural genomics pipelines, specifically those targeting families containing multiple prokaryotic orthologues. In measuring the domain coverage of the genomes, we show the benefits of selecting targets from both structurally uncharacterized domain families, whilst in addition, pursuing additional targets from large structurally characterised protein superfamilies.
Conclusion: This work suggests that such a combined approach to target selection is essential if structural genomics is to achieve a comprehensive structural coverage of the genomes, leading to greater insights into structure and the mechanisms that underlie protein evolution.
Figures



Similar articles
-
PSI-2: structural genomics to cover protein domain family space.Structure. 2009 Jun 10;17(6):869-81. doi: 10.1016/j.str.2009.03.015. Structure. 2009. PMID: 19523904 Free PMC article. Review.
-
Progress of structural genomics initiatives: an analysis of solved target structures.J Mol Biol. 2005 May 20;348(5):1235-60. doi: 10.1016/j.jmb.2005.03.037. Epub 2005 Apr 2. J Mol Biol. 2005. PMID: 15854658
-
Comprehensive genome analysis of 203 genomes provides structural genomics with new insights into protein family space.Nucleic Acids Res. 2006 Feb 15;34(3):1066-80. doi: 10.1093/nar/gkj494. Print 2006. Nucleic Acids Res. 2006. PMID: 16481312 Free PMC article.
-
SUPFAM--a database of potential protein superfamily relationships derived by comparing sequence-based and structure-based families: implications for structural genomics and function annotation in genomes.Nucleic Acids Res. 2002 Jan 1;30(1):289-93. doi: 10.1093/nar/30.1.289. Nucleic Acids Res. 2002. PMID: 11752317 Free PMC article.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
-
fDETECT webserver: fast predictor of propensity for protein production, purification, and crystallization.BMC Bioinformatics. 2018 Jan 3;18(1):580. doi: 10.1186/s12859-017-1995-z. BMC Bioinformatics. 2018. PMID: 29295714 Free PMC article.
-
PSI-2: structural genomics to cover protein domain family space.Structure. 2009 Jun 10;17(6):869-81. doi: 10.1016/j.str.2009.03.015. Structure. 2009. PMID: 19523904 Free PMC article. Review.
-
The Jpred 3 secondary structure prediction server.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W197-201. doi: 10.1093/nar/gkn238. Epub 2008 May 7. Nucleic Acids Res. 2008. PMID: 18463136 Free PMC article.
-
Unexpected features of the dark proteome.Proc Natl Acad Sci U S A. 2015 Dec 29;112(52):15898-903. doi: 10.1073/pnas.1508380112. Epub 2015 Nov 17. Proc Natl Acad Sci U S A. 2015. PMID: 26578815 Free PMC article.
-
Protein-folding chaperones predict structure-function relationships and cancer risk in BRCA1 mutation carriers.Cell Rep. 2024 Feb 27;43(2):113803. doi: 10.1016/j.celrep.2024.113803. Epub 2024 Feb 17. Cell Rep. 2024. PMID: 38368609 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

