Mapping and restriction of a dominant viral CD4+ T cell core epitope by both MHC class I and MHC class II
- PMID: 17320138
- PMCID: PMC1976554
- DOI: 10.1016/j.virol.2006.12.025
Mapping and restriction of a dominant viral CD4+ T cell core epitope by both MHC class I and MHC class II
Abstract
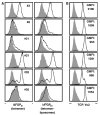
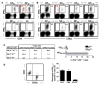
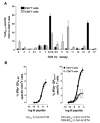
Virus-specific CD4(+) T cells contribute to effective virus control through a multiplicity of mechanisms including direct effector functions as well as "help" for B cell and CD8(+) T cell responses. Here, we have used the lymphocytic choriomeningitis virus (LCMV) system to assess the minimal constraints of a dominant antiviral CD4(+) T cell response. We report that the core epitope derived from the LCMV glycoprotein (GP) is 11 amino acids in length and provides optimal recognition by epitope-specific CD4(+) T cells. Surprisingly, this epitope is also recognized by LCMV-specific CD8(+) T cells and thus constitutes a unique viral determinant with dual MHC class I- and II-restriction.
Figures
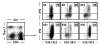


Similar articles
-
Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection.Eur J Immunol. 1998 Jan;28(1):390-400. doi: 10.1002/(SICI)1521-4141(199801)28:01<390::AID-IMMU390>3.0.CO;2-O. Eur J Immunol. 1998. PMID: 9485218
-
Conversion of tyrosine to the inflammation-associated analog 3'-nitrotyrosine at either TCR- or MHC-contact positions can profoundly affect recognition of the MHC class I-restricted epitope of lymphocytic choriomeningitis virus glycoprotein 33 by CD8 T cells.J Immunol. 2008 May 1;180(9):5956-62. doi: 10.4049/jimmunol.180.9.5956. J Immunol. 2008. PMID: 18424715
-
The signal sequence of lymphocytic choriomeningitis virus contains an immunodominant cytotoxic T cell epitope that is restricted by both H-2D(b) and H-2K(b) molecules.Virology. 1997 Jul 21;234(1):62-73. doi: 10.1006/viro.1997.8627. Virology. 1997. PMID: 9234947
-
Identification of MHC class II-restricted tumor antigens recognized by CD4+ T cells.Methods. 2003 Mar;29(3):227-35. doi: 10.1016/s1046-2023(02)00345-6. Methods. 2003. PMID: 12725788 Review.
-
Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes.Methods. 2003 Mar;29(3):282-8. doi: 10.1016/s1046-2023(02)00350-x. Methods. 2003. PMID: 12725793 Review.
Cited by
-
The protein tyrosine phosphatase PTPN22 controls forkhead box protein 3 T regulatory cell induction but is dispensable for T helper type 1 cell polarization.Clin Exp Immunol. 2014 Oct;178(1):178-89. doi: 10.1111/cei.12393. Clin Exp Immunol. 2014. PMID: 24905474 Free PMC article.
-
High efficiency of antiviral CD4(+) killer T cells.PLoS One. 2013;8(4):e60420. doi: 10.1371/journal.pone.0060420. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565245 Free PMC article.
-
Antigen-specific CD4 T-cell help rescues exhausted CD8 T cells during chronic viral infection.Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21182-7. doi: 10.1073/pnas.1118450109. Epub 2011 Dec 12. Proc Natl Acad Sci U S A. 2011. PMID: 22160724 Free PMC article.
-
Chemokine Signatures of Pathogen-Specific T Cells I: Effector T Cells.J Immunol. 2020 Oct 15;205(8):2169-2187. doi: 10.4049/jimmunol.2000253. Epub 2020 Sep 18. J Immunol. 2020. PMID: 32948687 Free PMC article.
-
Type I IFNs and CD8 T cells increase intestinal barrier permeability after chronic viral infection.J Exp Med. 2020 Dec 7;217(12):e20192276. doi: 10.1084/jem.20192276. J Exp Med. 2020. PMID: 32880630 Free PMC article.
References
-
- Apostolopoulos V, Yu M, Corper AL, Li W, McKenzie IF, Teyton L, Wilson IA, Plebanski M. Crystal structure of a non-canonical high affinity peptide complexed with MHC class I: a novel use of alternative anchors. J Mol Biol. 2002;318(5):1307–1316. - PubMed
-
- Belkaid Y, Rouse BT. Natural regulatory T cells in infectious disease. Nat Immunol. 2005;6(4):353–360. - PubMed
-
- Berger DP, Homann D, Oldstone MB. Defining parameters for successful immunocytotherapy of persistent viral infection. Virology. 2000;266(2):257–263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

