A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere
- PMID: 17318187
- PMCID: PMC1817632
- DOI: 10.1038/sj.emboj.7601584
A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere
Abstract
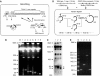
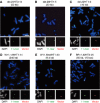
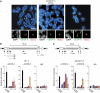
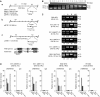
Chromatin clusters containing CENP-A, a histone H3 variant, are found in centromeres of multicellular eukaryotes. This study examines the ability of alpha-satellite (alphoid) DNA arrays in different lengths to nucleate CENP-A chromatin and form functional kinetochores de novo. Kinetochore assembly was followed by measuring human artificial chromosome formation in cultured human cells and by chromatin immunoprecipitation analysis. The results showed that both the length of alphoid DNA arrays and the density of CENP-B boxes had a strong impact on nucleation, spreading and/or maintenance of CENP-A chromatin, and formation of functional kinetochores. These effects are attributed to a change in the dynamic balance between assembly of chromatin containing trimethyl histone H3-K9 and chromatin containing CENP-A/C. The data presented here suggest that a functional minimum core stably maintained on 30-70 kb alphoid DNA arrays represents an epigenetic memory of centromeric chromatin.
Figures


Similar articles
-
CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA.J Cell Biol. 2002 Dec 9;159(5):765-75. doi: 10.1083/jcb.200207112. Epub 2002 Dec 2. J Cell Biol. 2002. PMID: 12460987 Free PMC article.
-
Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes.J Cell Sci. 2003 Oct 1;116(Pt 19):4021-34. doi: 10.1242/jcs.00697. J Cell Sci. 2003. PMID: 12953060
-
Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere.Hum Mol Genet. 2005 Jan 1;14(1):85-93. doi: 10.1093/hmg/ddi008. Epub 2004 Nov 10. Hum Mol Genet. 2005. PMID: 15537667
-
How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly.Biochem Cell Biol. 2006 Aug;84(4):619-39. doi: 10.1139/o06-078. Biochem Cell Biol. 2006. PMID: 16936833 Review.
-
Heterochromatin tells CENP-A where to go.Bioessays. 2008 Jun;30(6):526-9. doi: 10.1002/bies.20763. Bioessays. 2008. PMID: 18478529 Review.
Cited by
-
Hypermorphic expression of centromeric retroelement-encoded small RNAs impairs CENP-A loading.Chromosome Res. 2013 Mar;21(1):49-62. doi: 10.1007/s10577-013-9337-0. Epub 2013 Feb 8. Chromosome Res. 2013. PMID: 23392618
-
DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function.Dev Cell. 2015 May 4;33(3):314-27. doi: 10.1016/j.devcel.2015.03.020. Dev Cell. 2015. PMID: 25942623 Free PMC article.
-
Centromeres under Pressure: Evolutionary Innovation in Conflict with Conserved Function.Genes (Basel). 2020 Aug 10;11(8):912. doi: 10.3390/genes11080912. Genes (Basel). 2020. PMID: 32784998 Free PMC article. Review.
-
Genetic and epigenetic regulation of centromeres: a look at HAC formation.Chromosome Res. 2015 Feb;23(1):87-103. doi: 10.1007/s10577-015-9470-z. Chromosome Res. 2015. PMID: 25682171 Review.
-
Transposable elements: genome innovation, chromosome diversity, and centromere conflict.Chromosome Res. 2018 Mar;26(1-2):5-23. doi: 10.1007/s10577-017-9569-5. Epub 2018 Jan 13. Chromosome Res. 2018. PMID: 29332159 Free PMC article. Review.
References
-
- Amor DJ, Kalitsis P, Sumer H, Choo KH (2004) Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol 14: 359–368 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

