Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1
- PMID: 17306567
- PMCID: PMC1885958
- DOI: 10.1016/j.immuni.2007.01.007
Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1
Abstract
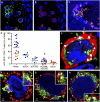
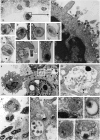
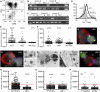
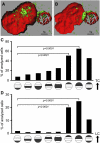
Understanding the initial events in the establishment of vaginal human immunodeficiency virus type-1 (HIV-1) entry and infection has been hampered by the lack of appropriate experimental models. Here, we show in an ex vivo human organ culture system that upon contact in situ, HIV-1 rapidly penetrated both intraepithelial vaginal Langerhans and CD4(+) T cells. HIV-1 entered CD4(+) T cells almost exclusively by CD4 and CCR5 receptor-mediated direct fusion, without requiring passage from Langerhans cells, and overt productive infection ensued. By contrast, HIV-1 entered CD1a(+) Langerhans cells primarily by endocytosis, by means of multiple receptors, and virions persisted intact within the cytoplasm for several days. Our findings shed light on the very earliest steps of mucosal HIV infection in vivo and may guide the design of effective strategies to block local transmission and prevent HIV-1 spread.
Figures


Comment in
-
HIV's vagina travelogue.Immunity. 2007 Feb;26(2):145-7. doi: 10.1016/j.immuni.2007.02.001. Immunity. 2007. PMID: 17307703 Review.
-
HIV transmission: migratory Langerhans cells are primary targets in vaginal HIV transmission.Immunol Cell Biol. 2007 Jun;85(4):269-70. doi: 10.1038/sj.icb.7100058. Epub 2007 Apr 10. Immunol Cell Biol. 2007. PMID: 17420767 No abstract available.
Similar articles
-
HIV's vagina travelogue.Immunity. 2007 Feb;26(2):145-7. doi: 10.1016/j.immuni.2007.02.001. Immunity. 2007. PMID: 17307703 Review.
-
Vaginal langerhans cells nonproductively transporting HIV-1 mediate infection of T cells.J Virol. 2011 Dec;85(24):13443-7. doi: 10.1128/JVI.05615-11. Epub 2011 Oct 5. J Virol. 2011. PMID: 21976645 Free PMC article.
-
Early events in HIV transmission through a human reconstructed vaginal mucosa.AIDS. 2008 Jul 11;22(11):1257-66. doi: 10.1097/QAD.0b013e3282f736f4. AIDS. 2008. PMID: 18580604
-
Transmission, acute HIV-1 infection and the quest for strategies to prevent infection.Nat Med. 2003 Jul;9(7):847-52. doi: 10.1038/nm0703-847. Nat Med. 2003. PMID: 12835704 Review.
-
In vivo identification of Langerhans and related dendritic cells infected with HIV-1 subtype E in vaginal mucosa of asymptomatic patients.Mod Pathol. 2001 Dec;14(12):1263-9. doi: 10.1038/modpathol.3880472. Mod Pathol. 2001. PMID: 11743049
Cited by
-
The importance of the vaginal delivery route for antiretrovirals in HIV prevention.Ther Deliv. 2011 Dec;2(12):1535-50. doi: 10.4155/tde.11.126. Ther Deliv. 2011. PMID: 22468220 Free PMC article. Review.
-
Mucosal effects of tenofovir 1% gel.Elife. 2015 Feb 3;4:e04525. doi: 10.7554/eLife.04525. Elife. 2015. PMID: 25647729 Free PMC article. Clinical Trial.
-
Cell-to-Cell Spreading of HIV-1 in Myeloid Target Cells Escapes SAMHD1 Restriction.mBio. 2019 Nov 19;10(6):e02457-19. doi: 10.1128/mBio.02457-19. mBio. 2019. PMID: 31744918 Free PMC article.
-
HIV-1 replicates and persists in vaginal epithelial dendritic cells.J Clin Invest. 2018 Aug 1;128(8):3439-3444. doi: 10.1172/JCI98943. Epub 2018 Jul 9. J Clin Invest. 2018. PMID: 29723162 Free PMC article. Clinical Trial.
-
Defining the interaction of HIV-1 with the mucosal barriers of the female reproductive tract.J Virol. 2013 Nov;87(21):11388-400. doi: 10.1128/JVI.01377-13. Epub 2013 Aug 21. J Virol. 2013. PMID: 23966398 Free PMC article.
References
-
- Collins K.B., Patterson B.K., Naus G.J., Landers D.V., Gupta P. Development of an in vitro organ culture model to study transmission of HIV-1 in the female genital tract. Nat. Med. 2000;6:475–479. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

