Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis
- PMID: 17261847
- PMCID: PMC2063955
- DOI: 10.1083/jcb.200604141
Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis
Abstract
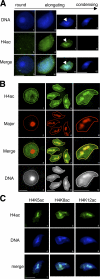
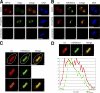
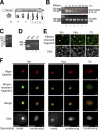
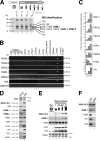
During male germ cell postmeiotic maturation, dramatic chromatin reorganization occurs, which is driven by completely unknown mechanisms. For the first time, we describe a specific reprogramming of mouse pericentric heterochromatin. Initiated when histones undergo global acetylation in early elongating spermatids, this process leads to the establishment of new DNA packaging structures organizing the pericentric regions in condensing spermatids. Five new histone variants were discovered, which are expressed in late spermiogenic cells. Two of them, which we named H2AL1 and H2AL2, specifically mark the pericentric regions in condensing spermatids and participate in the formation of new nucleoprotein structures. Moreover, our investigations also suggest that TH2B, an already identified testis-specific H2B variant of unknown function, could provide a platform for the structural transitions accompanying the incorporation of these new histone variants.
Figures


Similar articles
-
[Spermiogenesis: histone acetylation triggers male genome reprogramming].Gynecol Obstet Fertil. 2009 Jun;37(6):519-22. doi: 10.1016/j.gyobfe.2009.04.003. Epub 2009 May 17. Gynecol Obstet Fertil. 2009. PMID: 19447664 French.
-
Increased accessibility of the N-terminus of testis-specific histone TH2B to antibodies in elongating spermatids.Mol Reprod Dev. 1995 Oct;42(2):210-9. doi: 10.1002/mrd.1080420210. Mol Reprod Dev. 1995. PMID: 8562066
-
Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases.Eur J Cell Biol. 2000 Dec;79(12):950-60. doi: 10.1078/0171-9335-00123. Eur J Cell Biol. 2000. PMID: 11152286
-
A haploid affair: core histone transitions during spermatogenesis.Biochem Cell Biol. 2003 Jun;81(3):131-40. doi: 10.1139/o03-045. Biochem Cell Biol. 2003. PMID: 12897846 Review.
-
From meiosis to postmeiotic events: the secrets of histone disappearance.FEBS J. 2010 Feb;277(3):599-604. doi: 10.1111/j.1742-4658.2009.07504.x. Epub 2009 Dec 15. FEBS J. 2010. PMID: 20015078 Review.
Cited by
-
Structure, dynamics, and evolution of centromeric nucleosomes.Proc Natl Acad Sci U S A. 2007 Oct 9;104(41):15974-81. doi: 10.1073/pnas.0707648104. Epub 2007 Sep 24. Proc Natl Acad Sci U S A. 2007. PMID: 17893333 Free PMC article. Review.
-
1q12 chromosome translocations form aberrant heterochromatic foci associated with changes in nuclear architecture and gene expression in B cell lymphoma.EMBO Mol Med. 2010 May;2(5):159-71. doi: 10.1002/emmm.201000067. EMBO Mol Med. 2010. PMID: 20432501 Free PMC article.
-
Every amino acid matters: essential contributions of histone variants to mammalian development and disease.Nat Rev Genet. 2014 Apr;15(4):259-71. doi: 10.1038/nrg3673. Epub 2014 Mar 11. Nat Rev Genet. 2014. PMID: 24614311 Free PMC article. Review.
-
Dpy19l2-deficient globozoospermic sperm display altered genome packaging and DNA damage that compromises the initiation of embryo development.Mol Hum Reprod. 2015 Feb;21(2):169-85. doi: 10.1093/molehr/gau099. Epub 2014 Oct 29. Mol Hum Reprod. 2015. PMID: 25354700 Free PMC article.
-
SLY regulates genes involved in chromatin remodeling and interacts with TBL1XR1 during sperm differentiation.Cell Death Differ. 2017 Jun;24(6):1029-1044. doi: 10.1038/cdd.2017.32. Epub 2017 May 5. Cell Death Differ. 2017. PMID: 28475176 Free PMC article.
References
-
- Aul, R.B., and R.J. Oko. 2001. The major subacrosomal occupant of bull spermatozoa is a novel histone H2B variant associated with the forming acrosome during spermiogenesis. Dev. Biol. 239:376–387. - PubMed
-
- Caron, C., J. Govin, S. Rousseaux, and S. Khochbin. 2005. How to pack the genome for a safe trip. Prog. Mol. Subcell. Biol. 38:65–89. - PubMed
-
- Churikov, D., J. Siino, M. Svetlova, K. Zhang, A. Gineitis, E. Morton Bradbury, and A. Zalensky. 2004. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics. 84:745–756. - PubMed
-
- Faure, A.K., C. Pivot-Pajot, A. Kerjean, M. Hazzouri, R. Pelletier, M. Peoc'h, B. Sele, S. Khochbin, and S. Rousseaux. 2003. Misregulation of histone acetylation in Sertoli cell-only syndrome and testicular cancer. Mol. Hum. Reprod. 9:757–763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

