Conservation of noncoding microsatellites in plants: implication for gene regulation
- PMID: 17187690
- PMCID: PMC1781443
- DOI: 10.1186/1471-2164-7-323
Conservation of noncoding microsatellites in plants: implication for gene regulation
Abstract
Background: Microsatellites are extremely common in plant genomes, and in particular, they are significantly enriched in the 5' noncoding regions. Although some 5' noncoding microsatellites involved in gene regulation have been described, the general properties of microsatellites as regulatory elements are still unknown. To address the question of microsatellites associated with regulatory elements, we have analyzed the conserved noncoding microsatellite sequences (CNMSs) in the 5' noncoding regions by inter- and intragenomic phylogenetic footprinting in the Arabidopsis and Brassica genomes.
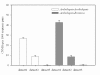
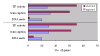
Results: We identified 247 Arabidopsis-Brassica orthologous and 122 Arabidopsis paralogous CNMSs, representing 491 CT/GA and CTT/GAA repeats, which accounted for 10.6% of these types located in the 500-bp regions upstream of coding sequences in the Arabidopsis genome. Among these identified CNMSs, 18 microsatellites show high conservation in the regulatory regions of both orthologous and paralogous genes, and some of them also appear in the corresponding positions of more distant homologs in Arabidopsis, as well as in other plants. A computational scan of CNMSs for known cis-regulatory elements showed that light responsive elements were clustered in the region of CT/GA repeats, as well as salicylic acid responsive elements in the (CTT)n/(GAA)n sequences. Patterns of gene expression revealed that 70-80% of CNMS (CTT)n/(GAA)n associated genes were regulated by salicylic acid, which was consistent with the prediction of regulatory elements in silico.
Conclusion: Our analyses showed that some noncoding microsatellites were conserved in plants and appeared to be ancient. These CNMSs served as regulatory elements involved in light and salicylic acid responses. Our findings might have implications in the common features of the over-represented microsatellites for gene regulation in plant-specific pathways.
Figures
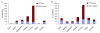






Similar articles
-
Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice.BMC Genomics. 2007 Aug 1;8:260. doi: 10.1186/1471-2164-8-260. BMC Genomics. 2007. PMID: 17672917 Free PMC article.
-
Microsatellites in Brassica unigenes: relative abundance, marker design, and use in comparative physical mapping and genome analysis.Genome. 2010 Jan;53(1):55-67. doi: 10.1139/g09-084. Genome. 2010. PMID: 20130749
-
Comparative analysis of noncoding sequences of orthologous bovine and human gene pairs.Genet Mol Res. 2004 Dec 30;3(4):465-73. Genet Mol Res. 2004. PMID: 15688313
-
The evolution of plant regulatory networks: what Arabidopsis cannot say for itself.Curr Opin Plant Biol. 2007 Dec;10(6):653-9. doi: 10.1016/j.pbi.2007.07.009. Epub 2007 Aug 27. Curr Opin Plant Biol. 2007. PMID: 17720614 Review.
-
Promoter microsatellites as modulators of human gene expression.Adv Exp Med Biol. 2012;769:41-54. doi: 10.1007/978-1-4614-5434-2_4. Adv Exp Med Biol. 2012. PMID: 23560304 Review.
Cited by
-
Genome-wide cis-regulatory signatures for modulation of agronomic traits as exemplified by drought yield index (DYI) in chickpea.Funct Integr Genomics. 2019 Nov;19(6):973-992. doi: 10.1007/s10142-019-00691-2. Epub 2019 Jun 8. Funct Integr Genomics. 2019. PMID: 31177403
-
Tandem repeat distribution of gene transcripts in three plant families.Genet Mol Biol. 2009 Oct;32(4):822-33. doi: 10.1590/S1415-47572009005000091. Epub 2009 Dec 1. Genet Mol Biol. 2009. PMID: 21637460 Free PMC article.
-
An analysis of the Athetis lepigone transcriptome from four developmental stages.PLoS One. 2013 Sep 13;8(9):e73911. doi: 10.1371/journal.pone.0073911. eCollection 2013. PLoS One. 2013. PMID: 24058501 Free PMC article.
-
Development of genome-wide informative simple sequence repeat markers for large-scale genotyping applications in chickpea and development of web resource.Front Plant Sci. 2015 Aug 21;6:645. doi: 10.3389/fpls.2015.00645. eCollection 2015. Front Plant Sci. 2015. PMID: 26347762 Free PMC article.
-
A polymorphic (GA/CT)n- SSR influences promoter activity of Tryptophan decarboxylase gene in Catharanthus roseus L. Don.Sci Rep. 2016 Sep 13;6:33280. doi: 10.1038/srep33280. Sci Rep. 2016. PMID: 27623355 Free PMC article.
References
-
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. - PubMed
-
- Fujimori S, Washio T, Higo K, Ohtomo Y, Murakami K, Matsubara K, Kawai J, Carninci P, Hayashizaki Y, Kikuchi S, Tomita M. A novel feature of microsatellites in plants: a distribution gradient along the direction of transcription. FEBS Lett. 2003;554:17–22. doi: 10.1016/S0014-5793(03)01041-X. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

