Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast
- PMID: 17179073
- PMCID: PMC1840069
- DOI: 10.1534/genetics.106.064170
Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast
Abstract
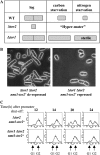
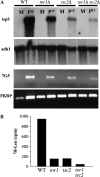
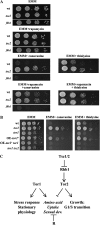
The TOR protein kinases exhibit a conserved role in regulating cellular growth and proliferation. In the fission yeast two TOR homologs are present. tor1(+) is required for starvation and stress responses, while tor2(+) is essential. We report here that Tor2 depleted cells show a phenotype very similar to that of wild-type cells starved for nitrogen, including arrest at the G(1) phase of the cell cycle, induction of nitrogen-starvation-specific genes, and entrance into the sexual development pathway. The phenotype of tor2 mutants is in a striking contrast to the failure of tor1 mutants to initiate sexual development or arrest in G(1) under nitrogen starvation conditions. Tsc1 and Tsc2, the genes mutated in the human tuberous sclerosis complex syndrome, negatively regulate the mammalian TOR via inactivation of the GTPase Rheb. We analyzed the genetic relationship between the two TOR genes and the Schizosaccharomyces pombe orthologs of TSC1, TSC2, and Rheb. Our data suggest that like in higher eukaryotes, the Tsc1-2 complex negatively regulates Tor2. In contrast, the Tsc1-2 complex and Tor1 appear to work in parallel, both positively regulating amino acid uptake through the control of expression of amino acid permeases. Additionally, either Tsc1/2 or Tor1 are required for growth on a poor nitrogen source such as proline. Mutants lacking Tsc1 or Tsc2 are highly sensitive to rapamycin under poor nitrogen conditions, suggesting that the function of Tor1 under such conditions is sensitive to rapamycin. We discuss the complex genetic interactions between tor1(+), tor2(+), and tsc1/2(+) and the implications for rapamycin sensitivity in tsc1 or tsc2 mutants.
Figures


Similar articles
-
Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin.Genetics. 2005 Feb;169(2):539-50. doi: 10.1534/genetics.104.034983. Epub 2004 Sep 30. Genetics. 2005. PMID: 15466417 Free PMC article.
-
Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase.Genes Cells. 2006 Dec;11(12):1367-79. doi: 10.1111/j.1365-2443.2006.01025.x. Genes Cells. 2006. PMID: 17121544
-
Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast.Mol Cell Biol. 2007 Apr;27(8):3154-64. doi: 10.1128/MCB.01039-06. Epub 2007 Jan 29. Mol Cell Biol. 2007. PMID: 17261596 Free PMC article.
-
Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase.Oncogene. 2006 Oct 16;25(48):6361-72. doi: 10.1038/sj.onc.1209882. Oncogene. 2006. PMID: 17041622 Review.
-
TOR signaling in fission yeast.Crit Rev Biochem Mol Biol. 2008 Jul-Aug;43(4):277-83. doi: 10.1080/10409230802254911. Crit Rev Biochem Mol Biol. 2008. PMID: 18756382 Review.
Cited by
-
Altered nuclear tRNA metabolism in La-deleted Schizosaccharomyces pombe is accompanied by a nutritional stress response involving Atf1p and Pcr1p that is suppressible by Xpo-t/Los1p.Mol Biol Cell. 2012 Feb;23(3):480-91. doi: 10.1091/mbc.E11-08-0732. Epub 2011 Dec 7. Mol Biol Cell. 2012. PMID: 22160596 Free PMC article.
-
Fission yeast TOR signaling is essential for the down-regulation of a hyperactivated stress-response MAP kinase under salt stress.Mol Genet Genomics. 2013 Feb;288(1-2):63-75. doi: 10.1007/s00438-012-0731-7. Epub 2012 Dec 28. Mol Genet Genomics. 2013. PMID: 23271606
-
Fission Yeast TORC1 Promotes Cell Proliferation through Sfp1, a Transcription Factor Involved in Ribosome Biogenesis.Mol Cell Biol. 2023;43(12):675-692. doi: 10.1080/10985549.2023.2282349. Epub 2023 Dec 20. Mol Cell Biol. 2023. PMID: 38051102 Free PMC article.
-
TOR and PKA pathways synergize at the level of the Ste11 transcription factor to prevent mating and meiosis in fission yeast.PLoS One. 2010 Jul 9;5(7):e11514. doi: 10.1371/journal.pone.0011514. PLoS One. 2010. PMID: 20634885 Free PMC article.
-
Characterization of genome-reduced fission yeast strains.Nucleic Acids Res. 2013 May 1;41(10):5382-99. doi: 10.1093/nar/gkt233. Epub 2013 Apr 5. Nucleic Acids Res. 2013. PMID: 23563150 Free PMC article.
References
-
- Alvarez, B., and S. Moreno, 2006. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 119: 4475–4485. - PubMed
-
- Beck, T., and M. N. Hall, 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. - PubMed
-
- Di Como, C. J., and K. T. Arndt, 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10: 1904–1916. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

