Comparative analysis of hepatitis C virus phylogenies from coding and non-coding regions: the 5' untranslated region (UTR) fails to classify subtypes
- PMID: 17169155
- PMCID: PMC1764733
- DOI: 10.1186/1743-422X-3-103
Comparative analysis of hepatitis C virus phylogenies from coding and non-coding regions: the 5' untranslated region (UTR) fails to classify subtypes
Abstract
Background: The duration of treatment for HCV infection is partly indicated by the genotype of the virus. For studies of disease transmission, vaccine design, and surveillance for novel variants, subtype-level classification is also needed. This study used the Shimodaira-Hasegawa test and related statistical techniques to compare phylogenetic trees obtained from coding and non-coding regions of a whole-genome alignment for the reliability of subtyping in different regions.
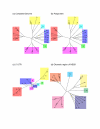
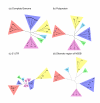
Results: Different regions of the HCV genome yield inconsistent phylogenies, which can lead to erroneous conclusions about classification of a given infection. In particular, the highly conserved 5' untranslated region (UTR) yields phylogenetic trees with topologies that differ from the HCV polyprotein and complete genome phylogenies. Phylogenetic trees from the NS5B gene reliably cluster related subtypes, and yield topologies consistent with those of the whole genome and polyprotein.
Conclusion: These results extend those from previous studies and indicate that, unlike the NS5B gene, the 5' UTR contains insufficient variation to resolve HCV classifications to the level of viral subtype, and fails to distinguish genotypes reliably. Use of the 5' UTR for clinical tests to characterize HCV infection should be replaced by a subtype-informative test.
Figures
Similar articles
-
Hepatitis C virus genotyping: interrogation of the 5' untranslated region cannot accurately distinguish genotypes 1a and 1b.J Clin Microbiol. 2002 Sep;40(9):3127-34. doi: 10.1128/JCM.40.9.3127-3134.2002. J Clin Microbiol. 2002. PMID: 12202542 Free PMC article.
-
Genotyping hepatitis C virus from hemodialysis patients in Central Brazil by line probe assay and sequence analysis.Braz J Med Biol Res. 2007 Apr;40(4):545-50. doi: 10.1590/s0100-879x2007000400013. Braz J Med Biol Res. 2007. PMID: 17401498
-
Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5' untranslated region sequences.J Clin Microbiol. 2007 Apr;45(4):1102-12. doi: 10.1128/JCM.02366-06. Epub 2007 Feb 7. J Clin Microbiol. 2007. PMID: 17287328 Free PMC article.
-
Update on different aspects of HCV variability: focus on NS5B polymerase.BMC Infect Dis. 2014;14 Suppl 5(Suppl 5):S1. doi: 10.1186/1471-2334-14-S5-S1. Epub 2014 Sep 5. BMC Infect Dis. 2014. PMID: 25234810 Free PMC article. Review.
-
[Initiation of genetic translation in HCV polyprotein].Nihon Rinsho. 2004 Jul;62 Suppl 7(Pt 1):48-53. Nihon Rinsho. 2004. PMID: 15359762 Review. Japanese. No abstract available.
Cited by
-
Analysis of genomic-length HBV sequences to determine genotype and subgenotype reference sequences.J Gen Virol. 2020 Mar;101(3):271-283. doi: 10.1099/jgv.0.001387. J Gen Virol. 2020. PMID: 32134374 Free PMC article.
-
Molecular epidemiology and putative origin of hepatitis C virus in random volunteers from Argentina.World J Gastroenterol. 2013 Sep 21;19(35):5813-27. doi: 10.3748/wjg.v19.i35.5813. World J Gastroenterol. 2013. PMID: 24124326 Free PMC article.
-
Hepatitis C Virus Subtypes Novel 6g-Related Subtype and 6w Could Be Indigenous in Southern Taiwan with Characteristic Geographic Distribution.Viruses. 2021 Jul 7;13(7):1316. doi: 10.3390/v13071316. Viruses. 2021. PMID: 34372521 Free PMC article.
-
Mixed Infections Unravel Novel HCV Inter-Genotypic Recombinant Forms within the Conserved IRES Region.Viruses. 2024 Apr 3;16(4):560. doi: 10.3390/v16040560. Viruses. 2024. PMID: 38675902 Free PMC article.
-
Classification of hepatitis C virus and human immunodeficiency virus-1 sequences with the branching index.J Gen Virol. 2008 Sep;89(Pt 9):2098-2107. doi: 10.1099/vir.0.83657-0. J Gen Virol. 2008. PMID: 18753218 Free PMC article.
References
-
- Hadziyannis SJ, Sette H, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H, Bernstein D, Rizzetto M, Zeuzem S, Pockros PJ, Lin A, Ackrill AM. Peginterferon-alpha 2a and ribavirin combination therapy in chronic hepatitis C - A randomized study of treatment duration and ribavirin dose . Ann Intern Med. 2004;140:346–355. - PubMed
-
- Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin-I T, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

