Interplay between chromatin and trans-acting factors on the IME2 promoter upon induction of the gene at the onset of meiosis
- PMID: 17158929
- PMCID: PMC1800723
- DOI: 10.1128/MCB.01661-06
Interplay between chromatin and trans-acting factors on the IME2 promoter upon induction of the gene at the onset of meiosis
Abstract
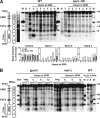
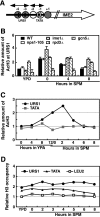
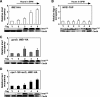
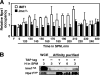
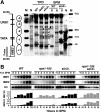
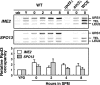
The IME2 gene is one of the key regulators of the initiation of meiosis in budding yeast. This gene is repressed during mitosis through the repressive chromatin structure at the promoter, which is maintained by the Rpd3-Sin3 histone deacetylase (HDAC) complex. IME2 expression in meiosis requires Gcn5/histone acetyltransferase, the transcriptional activator Ime1, and the chromatin remodeler RSC; however, the molecular basis of IME2 activation had not been previously defined. We found that, during mitotic growth, a nucleosome masked the TATA element of IME2, and this positioning depended on HDAC. This chromatin structure was remodeled at meiosis by RSC that was recruited to TATA by Ime1. Stable tethering of Ime1 to the promoter required the presence of Gcn5. Interestingly, Ime1 binding to the promoter was kept at low levels during the very early stages in meiosis, even when the levels of Ime1 and histone H3 acetylation at the promoter were at their highest, making a 4- to 6-h delay of the IME2 expression from that of IME1. HDAC was continuously present at the promoter regardless of the transcriptional condition of IME2, and deletion of RPD3 allowed the IME2 expression shortly after the expression of IME1, suggesting that HDAC plays a role in regulating the timing of IME2 expression.
Figures

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Comparison of cognitive behaviour therapy versus activity management, both delivered remotely, to treat paediatric chronic fatigue syndrome/myalgic encephalomyelitis: the UK FITNET-NHS RCT.Health Technol Assess. 2024 Oct;28(70):1-134. doi: 10.3310/VLRW6701. Health Technol Assess. 2024. PMID: 39485730 Free PMC article. Clinical Trial.
-
A multicomponent psychosocial intervention to reduce substance use by adolescents involved in the criminal justice system: the RISKIT-CJS RCT.Public Health Res (Southampt). 2023 Mar;11(3):1-77. doi: 10.3310/FKPY6814. Public Health Res (Southampt). 2023. PMID: 37254608
-
Topical fluoride as a cause of dental fluorosis in children.Cochrane Database Syst Rev. 2024 Jun 20;6(6):CD007693. doi: 10.1002/14651858.CD007693.pub3. Cochrane Database Syst Rev. 2024. PMID: 38899538 Review.
Cited by
-
RSC Chromatin-Remodeling Complex Is Important for Mitochondrial Function in Saccharomyces cerevisiae.PLoS One. 2015 Jun 18;10(6):e0130397. doi: 10.1371/journal.pone.0130397. eCollection 2015. PLoS One. 2015. PMID: 26086550 Free PMC article.
-
Yeast Rim11 kinase responds to glutathione-induced stress by regulating the transcription of phospholipid biosynthetic genes.Mol Biol Cell. 2024 Jan 1;35(1):ar8. doi: 10.1091/mbc.E23-03-0116. Epub 2023 Nov 8. Mol Biol Cell. 2024. PMID: 37938929 Free PMC article.
-
RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes.EMBO J. 2008 Jan 9;27(1):100-10. doi: 10.1038/sj.emboj.7601946. Epub 2007 Dec 6. EMBO J. 2008. PMID: 18059476 Free PMC article.
-
Ime1 and Ime2 are required for pseudohyphal growth of Saccharomyces cerevisiae on nonfermentable carbon sources.Mol Cell Biol. 2010 Dec;30(23):5514-30. doi: 10.1128/MCB.00390-10. Epub 2010 Sep 27. Mol Cell Biol. 2010. PMID: 20876298 Free PMC article.
-
The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 2012 Mar;76(1):1-15. doi: 10.1128/MMBR.05010-11. Microbiol Mol Biol Rev. 2012. PMID: 22390969 Free PMC article. Review.
References
-
- Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. - PubMed
-
- Burgess, S. M., M. Ajimura, and N. Kleckner. 1999. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc. Natl. Acad. Sci. USA 96:6835-6840. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
