Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway
- PMID: 17138701
- PMCID: PMC1693952
- DOI: 10.1105/tpc.105.036400
Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway
Abstract
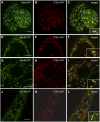
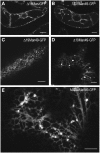
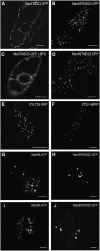
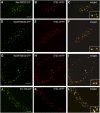
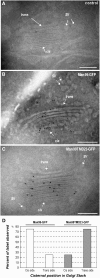
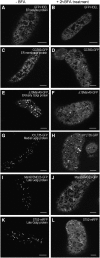
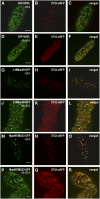
The processing of N-linked oligosaccharides in the secretory pathway requires the sequential action of a number of glycosidases and glycosyltransferases. We studied the spatial distribution of several type II membrane-bound enzymes from Glycine max, Arabidopsis thaliana, and Nicotiana tabacum. Glucosidase I (GCSI) localized to the endoplasmic reticulum (ER), alpha-1,2 mannosidase I (ManI) and N-acetylglucosaminyltransferase I (GNTI) both targeted to the ER and Golgi, and beta-1,2 xylosyltransferase localized exclusively to Golgi stacks, corresponding to the order of expected function. ManI deletion constructs revealed that the ManI transmembrane domain (TMD) contains all necessary targeting information. Likewise, GNTI truncations showed that this could apply to other type II enzymes. A green fluorescent protein chimera with ManI TMD, lengthened by duplicating its last seven amino acids, localized exclusively to the Golgi and colocalized with a trans-Golgi marker (ST52-mRFP), suggesting roles for protein-lipid interactions in ManI targeting. However, the TMD lengths of other plant glycosylation enzymes indicate that this mechanism cannot apply to all enzymes in the pathway. In fact, removal of the first 11 amino acids of the GCSI cytoplasmic tail resulted in relocalization from the ER to the Golgi, suggesting a targeting mechanism relying on protein-protein interactions. We conclude that the localization of N-glycan processing enzymes corresponds to an assembly line in the early secretory pathway and depends on both TMD length and signals in the cytoplasmic tail.
Figures





Similar articles
-
The Golgi Localization of GnTI Requires a Polar Amino Acid Residue within Its Transmembrane Domain.Plant Physiol. 2019 Jun;180(2):859-873. doi: 10.1104/pp.19.00310. Epub 2019 Apr 10. Plant Physiol. 2019. PMID: 30971450 Free PMC article.
-
Time-resolved fluorescence imaging reveals differential interactions of N-glycan processing enzymes across the Golgi stack in planta.Plant Physiol. 2013 Apr;161(4):1737-54. doi: 10.1104/pp.112.210757. Epub 2013 Feb 11. Plant Physiol. 2013. PMID: 23400704 Free PMC article.
-
Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera.Plant Cell. 2002 Jan;14(1):237-61. doi: 10.1105/tpc.010237. Plant Cell. 2002. PMID: 11826310 Free PMC article.
-
Multiple targets for brefeldin A.Cell. 1991 Nov 1;67(3):449-51. doi: 10.1016/0092-8674(91)90517-3. Cell. 1991. PMID: 1934055 Review. No abstract available.
-
Glucosidase II and MRH-domain containing proteins in the secretory pathway.Curr Protein Pept Sci. 2015;16(1):31-48. doi: 10.2174/1389203716666150213160438. Curr Protein Pept Sci. 2015. PMID: 25692846 Free PMC article. Review.
Cited by
-
A cascade of arabinosyltransferases controls shoot meristem size in tomato.Nat Genet. 2015 Jul;47(7):784-92. doi: 10.1038/ng.3309. Epub 2015 May 25. Nat Genet. 2015. PMID: 26005869
-
A small molecule inhibitor partitions two distinct pathways for trafficking of tonoplast intrinsic proteins in Arabidopsis.PLoS One. 2012;7(9):e44735. doi: 10.1371/journal.pone.0044735. Epub 2012 Sep 5. PLoS One. 2012. PMID: 22957103 Free PMC article.
-
Roles of N-terminal fatty acid acylations in membrane compartment partitioning: Arabidopsis h-type thioredoxins as a case study.Plant Cell. 2013 Mar;25(3):1056-77. doi: 10.1105/tpc.112.106849. Epub 2013 Mar 29. Plant Cell. 2013. PMID: 23543785 Free PMC article.
-
cis-Golgi proteins accumulate near the ER exit sites and act as the scaffold for Golgi regeneration after brefeldin A treatment in tobacco BY-2 cells.Mol Biol Cell. 2012 Aug;23(16):3203-14. doi: 10.1091/mbc.E12-01-0034. Epub 2012 Jun 27. Mol Biol Cell. 2012. PMID: 22740633 Free PMC article.
-
Organelle Visualization With Multicolored Fluorescent Markers in Bamboo.Front Plant Sci. 2021 Apr 15;12:658836. doi: 10.3389/fpls.2021.658836. eCollection 2021. Front Plant Sci. 2021. PMID: 33936145 Free PMC article.
References
-
- Andreeva, A.V., Kutuzov, M.A., Evans, D.E., and Hawes, C. (1998). The structure of the Golgi apparatus: A hundred year of questions. J. Exp. Bot. 49 1281–1291.
-
- Ben-Tekaya, H., Miura, K., Pepperkok, R., and Hauri, H.P. (2004). Live imaging of bidirectional traffic from the ERGIC. J. Cell Sci. 118 357–367. - PubMed
-
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15 441–447. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

