Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion
- PMID: 17134730
- PMCID: PMC7103323
- DOI: 10.1016/j.virol.2006.10.034
Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion
Abstract
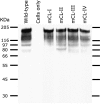
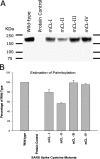
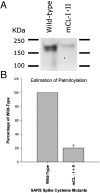
The SARS-coronavirus (SARS-CoV) is the etiological agent of the severe acute respiratory syndrome (SARS). The SARS-CoV spike (S) glycoprotein mediates membrane fusion events during virus entry and virus-induced cell-to-cell fusion. The cytoplasmic portion of the S glycoprotein contains four cysteine-rich amino acid clusters. Individual cysteine clusters were altered via cysteine-to-alanine amino acid replacement and the modified S glycoproteins were tested for their transport to cell-surfaces and ability to cause cell fusion in transient transfection assays. Mutagenesis of the cysteine cluster I, located immediately proximal to the predicted transmembrane, domain did not appreciably reduce cell-surface expression, although S-mediated cell fusion was reduced by more than 50% in comparison to the wild-type S. Similarly, mutagenesis of the cysteine cluster II located adjacent to cluster I reduced S-mediated cell fusion by more than 60% compared to the wild-type S, while cell-surface expression was reduced by less than 20%. Mutagenesis of cysteine clusters III and IV did not appreciably affect S cell-surface expression or S-mediated cell fusion. The wild-type S was palmitoylated as evidenced by the efficient incorporation of (3)H-palmitic acid in wild-type S molecules. S glycoprotein palmitoylation was significantly reduced for mutant glycoproteins having cluster I and II cysteine changes, but was largely unaffected for cysteine cluster III and IV mutants. These results show that the S cytoplasmic domain is palmitoylated and that palmitoylation of the membrane proximal cysteine clusters I and II may be important for S-mediated cell fusion.
Figures



Similar articles
-
Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion.Virology. 2005 Oct 25;341(2):215-30. doi: 10.1016/j.virol.2005.06.046. Epub 2005 Aug 15. Virology. 2005. PMID: 16099010 Free PMC article.
-
Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry.J Virol. 2006 Feb;80(3):1302-10. doi: 10.1128/JVI.80.3.1302-1310.2006. J Virol. 2006. PMID: 16415007 Free PMC article.
-
Mutagenesis of the transmembrane domain of the SARS coronavirus spike glycoprotein: refinement of the requirements for SARS coronavirus cell entry.Virol J. 2009 Dec 24;6:230. doi: 10.1186/1743-422X-6-230. Virol J. 2009. PMID: 20034394 Free PMC article.
-
The SARS-CoV S glycoprotein.Cell Mol Life Sci. 2004 Oct;61(19-20):2428-30. doi: 10.1007/s00018-004-4257-y. Cell Mol Life Sci. 2004. PMID: 15526150 Free PMC article. Review.
-
Site directed mutagenesis of the murine coronavirus spike protein. Effects on fusion.Adv Exp Med Biol. 1995;380:283-6. doi: 10.1007/978-1-4615-1899-0_45. Adv Exp Med Biol. 1995. PMID: 8830493 Review.
Cited by
-
Posttranslational modifications optimize the ability of SARS-CoV-2 spike for effective interaction with host cell receptors.Proc Natl Acad Sci U S A. 2022 Jul 12;119(28):e2119761119. doi: 10.1073/pnas.2119761119. Epub 2022 Jun 23. Proc Natl Acad Sci U S A. 2022. PMID: 35737823 Free PMC article.
-
Cell Entry of Animal Coronaviruses.Viruses. 2021 Oct 1;13(10):1977. doi: 10.3390/v13101977. Viruses. 2021. PMID: 34696406 Free PMC article. Review.
-
Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation.Nat Commun. 2021 Sep 9;12(1):5333. doi: 10.1038/s41467-021-25589-1. Nat Commun. 2021. PMID: 34504087 Free PMC article.
-
A graph-based approach identifies dynamic H-bond communication networks in spike protein S of SARS-CoV-2.J Struct Biol. 2020 Nov 1;212(2):107617. doi: 10.1016/j.jsb.2020.107617. Epub 2020 Sep 10. J Struct Biol. 2020. PMID: 32919067 Free PMC article.
-
Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein.Virology. 2010 Sep 15;405(1):139-48. doi: 10.1016/j.virol.2010.05.031. Epub 2010 Jul 1. Virology. 2010. PMID: 20580052 Free PMC article.
References
-
- Aiyar A., Xiang Y., Leis J. Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol. 1996;57:177–191. - PubMed
-
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. - PubMed
-
- Baker K.A., Dutch R.E., Lamb R.A., Jardetzky T.S. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3(3):309–319. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

