Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response
- PMID: 17132049
- PMCID: PMC1661684
- DOI: 10.1371/journal.pbio.0040423
Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response
Abstract
The protein folding capacity of the endoplasmic reticulum (ER) is regulated by the unfolded protein response (UPR). The UPR senses unfolded proteins in the ER lumen and transmits that information to the cell nucleus, where it drives a transcriptional program that is tailored to re-establish homeostasis. Using thin section electron microscopy, we found that yeast cells expand their ER volume at least 5-fold under UPR-inducing conditions. Surprisingly, we discovered that ER proliferation is accompanied by the formation of autophagosome-like structures that are densely and selectively packed with membrane stacks derived from the UPR-expanded ER. In analogy to pexophagy and mitophagy, which are autophagic processes that selectively sequester and degrade peroxisomes and mitochondria, the ER-specific autophagic process described utilizes several autophagy genes: they are induced by the UPR and are essential for the survival of cells subjected to severe ER stress. Intriguingly, cell survival does not require vacuolar proteases, indicating that ER sequestration into autophagosome-like structures, rather than their degradation, is the important step. Selective ER sequestration may help cells to maintain a new steady-state level of ER abundance even in the face of continuously accumulating unfolded proteins.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
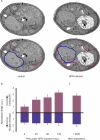
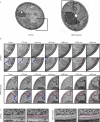

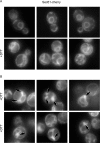
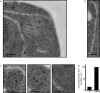

Comment in
-
Containing the damage of unfolded proteins.PLoS Biol. 2006 Dec;4(12):e442. doi: 10.1371/journal.pbio.0040442. Epub 2006 Nov 28. PLoS Biol. 2006. PMID: 20076517 Free PMC article. No abstract available.
Similar articles
-
ER-phagy: selective autophagy of the endoplasmic reticulum.Autophagy. 2007 May-Jun;3(3):285-7. doi: 10.4161/auto.3930. Epub 2007 May 1. Autophagy. 2007. PMID: 17351330
-
Endoplasmic reticulum stress triggers autophagy.J Biol Chem. 2006 Oct 6;281(40):30299-304. doi: 10.1074/jbc.M607007200. Epub 2006 Aug 10. J Biol Chem. 2006. PMID: 16901900 Free PMC article.
-
Increased autophagy reduces endoplasmic reticulum stress after neonatal hypoxia-ischemia: role of protein synthesis and autophagic pathways.Exp Neurol. 2014 May;255:103-12. doi: 10.1016/j.expneurol.2014.03.002. Epub 2014 Mar 12. Exp Neurol. 2014. PMID: 24631374
-
Pexophagy in yeasts.Biochim Biophys Acta. 2016 May;1863(5):992-8. doi: 10.1016/j.bbamcr.2015.09.023. Epub 2015 Sep 26. Biochim Biophys Acta. 2016. PMID: 26409485 Review.
-
Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium.Cell Death Differ. 2007 Sep;14(9):1576-82. doi: 10.1038/sj.cdd.4402200. Epub 2007 Jul 6. Cell Death Differ. 2007. PMID: 17612585 Review.
Cited by
-
Autophagic processes in yeast: mechanism, machinery and regulation.Genetics. 2013 Jun;194(2):341-61. doi: 10.1534/genetics.112.149013. Genetics. 2013. PMID: 23733851 Free PMC article. Review.
-
Autophagy: New Questions from Recent Answers.ISRN Mol Biol. 2012 Dec 30;2012:738718. doi: 10.5402/2012/738718. eCollection 2012. ISRN Mol Biol. 2012. PMID: 27335669 Free PMC article. Review.
-
Titanium Dioxide Nanoparticles Induce Endoplasmic Reticulum Stress-Mediated Autophagic Cell Death via Mitochondria-Associated Endoplasmic Reticulum Membrane Disruption in Normal Lung Cells.PLoS One. 2015 Jun 29;10(6):e0131208. doi: 10.1371/journal.pone.0131208. eCollection 2015. PLoS One. 2015. PMID: 26121477 Free PMC article.
-
ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load.J Cell Biol. 2012 Aug 6;198(3):371-85. doi: 10.1083/jcb.201202005. J Cell Biol. 2012. PMID: 22869598 Free PMC article.
-
Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart.Circ Res. 2015 Apr 10;116(8):1477-90. doi: 10.1161/CIRCRESAHA.116.303790. Circ Res. 2015. PMID: 25858070 Free PMC article. Review.
References
-
- Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. - PubMed
-
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. - PubMed
-
- McCracken AA, Brodsky JL. Recognition and delivery of ERAD substrates to the proteasome and alternative paths for cell survival. Curr Top Microbiol Immunol. 2005;300:17–40. - PubMed
-
- van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005;40:191–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

