Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination
- PMID: 17130289
- PMCID: PMC2064669
- DOI: 10.1083/jcb.200606145
Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination
Abstract
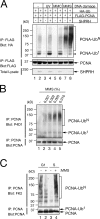
Differential modifications of proliferating cell nuclear antigen (PCNA) determine DNA repair pathways at stalled replication forks. In yeast, PCNA monoubiquitination by the ubiquitin ligase (E3) yRad18 promotes translesion synthesis (TLS), whereas the lysine-63-linked polyubiquitination of PCNA by yRad5 (E3) promotes the error-free mode of bypass. The yRad5-dependent pathway is important to prevent genomic instability during replication, although its exact molecular mechanism is poorly understood. This mechanism has remained totally elusive in mammals because of the lack of apparent RAD5 homologues. We report that a putative tumor suppressor gene, SHPRH, is a human orthologue of yeast RAD5. SHPRH associates with PCNA, RAD18, and the ubiquitin-conjugating enzyme UBC13 (E2) and promotes methyl methanesulfonate (MMS)-induced PCNA polyubiquitination. The reduction of SHPRH by stable short hairpin RNA increases sensitivity to MMS and enhances genomic instability. Therefore, the yRad5/SHPRH-dependent pathway is a conserved and fundamental DNA repair mechanism that protects the genome from genotoxic stress.
Figures



Similar articles
-
Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks.Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12411-6. doi: 10.1073/pnas.0805685105. Epub 2008 Aug 21. Proc Natl Acad Sci U S A. 2008. PMID: 18719106 Free PMC article.
-
Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen.Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18107-12. doi: 10.1073/pnas.0608595103. Epub 2006 Nov 15. Proc Natl Acad Sci U S A. 2006. PMID: 17108083 Free PMC article.
-
Synthesis of free and proliferating cell nuclear antigen-bound polyubiquitin chains by the RING E3 ubiquitin ligase Rad5.J Biol Chem. 2009 Oct 23;284(43):29326-34. doi: 10.1074/jbc.M109.043885. Epub 2009 Aug 25. J Biol Chem. 2009. PMID: 19706603 Free PMC article.
-
Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA.Mutat Res. 2017 Oct;803-805:82-88. doi: 10.1016/j.mrfmmm.2017.06.004. Epub 2017 Jun 21. Mutat Res. 2017. PMID: 28666590 Review.
-
Error-free DNA-damage tolerance in Saccharomyces cerevisiae.Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:43-50. doi: 10.1016/j.mrrev.2015.02.001. Epub 2015 Feb 16. Mutat Res Rev Mutat Res. 2015. PMID: 26041265 Review.
Cited by
-
Different types of interaction between PCNA and PIP boxes contribute to distinct cellular functions of Y-family DNA polymerases.Nucleic Acids Res. 2015 Sep 18;43(16):7898-910. doi: 10.1093/nar/gkv712. Epub 2015 Jul 13. Nucleic Acids Res. 2015. PMID: 26170230 Free PMC article.
-
Homologous recombination maintenance of genome integrity during DNA damage tolerance.Mol Cell Oncol. 2014 Oct 29;1(2):e957039. doi: 10.4161/23723548.2014.957039. eCollection 2014 Apr-Jun. Mol Cell Oncol. 2014. PMID: 27308329 Free PMC article. Review.
-
ncRNA-Encoded Peptides or Proteins and Cancer.Mol Ther. 2019 Oct 2;27(10):1718-1725. doi: 10.1016/j.ymthe.2019.09.001. Epub 2019 Sep 6. Mol Ther. 2019. PMID: 31526596 Free PMC article. Review.
-
Increased 26S proteasome non-ATPase regulatory subunit 1 in the aqueous humor of patients with age-related macular degeneration.BMB Rep. 2014 May;47(5):292-7. doi: 10.5483/bmbrep.2014.47.5.193. BMB Rep. 2014. PMID: 24286321 Free PMC article.
-
Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks.Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12411-6. doi: 10.1073/pnas.0805685105. Epub 2008 Aug 21. Proc Natl Acad Sci U S A. 2008. PMID: 18719106 Free PMC article.
References
-
- Friedberg, E.C., A.R. Lehmann, and R.P. Fuchs. 2005. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 18:499–505. - PubMed
-
- Hoege, C., B. Pfander, G.L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141. - PubMed
-
- Hofmann, R.M., and C.M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 96:645–653. - PubMed
-
- Huang, T.T., S.M. Nijman, K.D. Mirchandani, P.J. Galardy, M.A. Cohn, W. Haas, S.P. Gygi, H.L. Ploegh, R. Bernards, and A.D. D'Andrea. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8:339–347. - PubMed
-
- Johnson, R.E., S.T. Henderson, T.D. Petes, S. Prakash, M. Bankmann, and L. Prakash. 1992. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 12:3807–3818. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

