Human papillomavirus (HPV) types 101 and 103 isolated from cervicovaginal cells lack an E6 open reading frame (ORF) and are related to gamma-papillomaviruses
- PMID: 17125811
- PMCID: PMC1885239
- DOI: 10.1016/j.virol.2006.10.022
Human papillomavirus (HPV) types 101 and 103 isolated from cervicovaginal cells lack an E6 open reading frame (ORF) and are related to gamma-papillomaviruses
Abstract
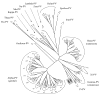
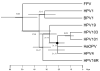
Complete genomes of HPV101 and HPV103 were PCR amplified and cloned from cervicovaginal cells of a 34-year-old female with cervical intraepithelial neoplasia grade 3 (CIN 3) and a 30-year-old female with a normal Pap test, respectively. HPV101 and HPV103 contain 4 early genes (E7, E1, E2, and E4) and 2 late genes (L2 and L1), but both lack the canonical E6 ORF. Pairwise alignment similarity of the L1 ORF nucleotide sequences of HPV101 and HPV103 indicated that they are at least 30% dissimilar to each other and all known PVs. However, similarities of the other ORFs (E7, E1, E2, and L2) indicated that HPV101 and HPV103 are most related to each other. Phylogenetic analyses revealed that these two types form a monophyletic clade, clustering together with the gamma- and pi-PV groups. These data demonstrated that HPV genomes closely related to papillomaviruses identified from cutaneous epithelia can be isolated from the genital mucosal region. Moreover, this is the first report of HPVs lacking an E6 ORF and phylogenetic evidence suggests this occurred subsequent to their emergence from the gamma-/pi-PVs.
Figures

Similar articles
-
Human papillomavirus 16 E6/E7 transcript and E2 gene status in patients with cervical neoplasia.Mol Diagn. 2004;8(1):57-64. doi: 10.1007/BF03260048. Mol Diagn. 2004. PMID: 15230643
-
Uniform distribution of HPV 16 E6 and E7 variants in patients with normal histology, cervical intra-epithelial neoplasia and cervical cancer.Int J Cancer. 1999 Jul 19;82(2):203-7. doi: 10.1002/(sici)1097-0215(19990719)82:2<203::aid-ijc9>3.0.co;2-9. Int J Cancer. 1999. PMID: 10389753
-
Identification of human papillomavirus type 53 L1, E6 and E7 variants in isolates from Brazilian women.Infect Genet Evol. 2012 Jan;12(1):71-6. doi: 10.1016/j.meegid.2011.10.018. Epub 2011 Oct 25. Infect Genet Evol. 2012. PMID: 22056883
-
Human papillomaviruses: genetic basis of carcinogenicity.Public Health Genomics. 2009;12(5-6):281-90. doi: 10.1159/000214919. Epub 2009 Aug 11. Public Health Genomics. 2009. PMID: 19684441 Free PMC article. Review.
-
[Virological and carcinogenic aspects of HPV].Bull Acad Natl Med. 2007 Mar;191(3):611-23; discussion 623. Bull Acad Natl Med. 2007. PMID: 18072657 Review. French.
Cited by
-
Papillomavirus E6 oncoproteins.Virology. 2013 Oct;445(1-2):115-37. doi: 10.1016/j.virol.2013.04.026. Epub 2013 May 24. Virology. 2013. PMID: 23711382 Free PMC article. Review.
-
Assessing Gammapapillomavirus infections of mucosal epithelia with two broad-spectrum PCR protocols.BMC Infect Dis. 2020 Apr 7;20(1):274. doi: 10.1186/s12879-020-4893-3. BMC Infect Dis. 2020. PMID: 32264841 Free PMC article.
-
Role of human papillomavirus infection in the etiology of vulvar cancer in Italian women.Infect Agent Cancer. 2020 Apr 1;15:20. doi: 10.1186/s13027-020-00286-8. eCollection 2020. Infect Agent Cancer. 2020. PMID: 32266002 Free PMC article.
-
The genetic diversity of human papillomavirus types from the species Gammapapillomavirus 15: HPV135, HPV146, and HPV179.PLoS One. 2021 May 6;16(5):e0249829. doi: 10.1371/journal.pone.0249829. eCollection 2021. PLoS One. 2021. PMID: 33956809 Free PMC article.
-
The biological properties of E6 and E7 oncoproteins from human papillomaviruses.Virus Genes. 2010 Feb;40(1):1-13. doi: 10.1007/s11262-009-0412-8. Epub 2009 Oct 17. Virus Genes. 2010. PMID: 19838783 Review.
References
-
- Andrews P. Evolution and environment in the Hominoidea. Nature. 1992;360(6405):641–6. - PubMed
-
- Antonsson A, Erfurt C, Hazard K, Holmgren V, Simon M, Kataoka A, Hossain S, Hakangard C, Hansson BG. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J Gen Virol. 2003;84(Pt 7):1881–6. - PubMed
-
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–52. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

