The promyelocytic leukemia protein functions as a negative regulator of IFN-gamma signaling
- PMID: 17121994
- PMCID: PMC1693728
- DOI: 10.1073/pnas.0604800103
The promyelocytic leukemia protein functions as a negative regulator of IFN-gamma signaling
Abstract
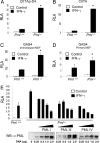
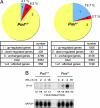
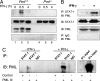
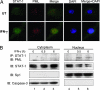
IFN-gamma is an immunomodulatory cytokine and uses the STAT-1alpha transcription factor to mediate gene expression. The promyelocytic leukemia (PML) protein regulates transcription as an activator or repressor, depending on the gene under investigation. Herein, we examined the influence of PML on IFN-gamma signaling, using PML wild-type (Pml(+/+)) and deficient (Pml(-/-)) mouse embryonic fibroblasts (MEF). Pml(-/-) MEF exhibit enhanced IFN-gamma-induced STAT-1alpha transcriptional activity compared with Pml(+/+) cells. Moreover, reconstitution of PML in Pml(-/-) MEF reduced STAT-1alpha transcriptional activity to levels comparable to Pml(+/+) MEF. Numerous endogenous IFN-gamma-regulated genes were up-regulated in Pml(-/-) MEF compared with Pml(+/+) MEF. IFN-gamma-mediated STAT-1alpha DNA-binding activity was enhanced in Pml(-/-) cells compared with Pml(+/+) cells. Lastly, IFN-gamma enhanced the formation of a PML-STAT-1alpha complex in the nucleus. These data suggest a novel function for PML in the IFN-gamma signaling pathway by inhibiting STAT-1alpha DNA binding and transcriptional activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
PML positively regulates interferon gamma signaling.Biochimie. 2011 Mar;93(3):389-98. doi: 10.1016/j.biochi.2010.11.005. Epub 2010 Nov 27. Biochimie. 2011. PMID: 21115099
-
Loss of the promyelocytic leukemia protein in gastric cancer: implications for IP-10 expression and tumor-infiltrating lymphocytes.PLoS One. 2011;6(10):e26264. doi: 10.1371/journal.pone.0026264. Epub 2011 Oct 12. PLoS One. 2011. PMID: 22022583 Free PMC article.
-
Involvement of promyelocytic leukemia protein in the ethanol-induced apoptosis in mouse embryo fibroblasts.Yakugaku Zasshi. 2008 Jul;128(7):1067-71. doi: 10.1248/yakushi.128.1067. Yakugaku Zasshi. 2008. PMID: 18591875
-
PML control of cytokine signaling.Cytokine Growth Factor Rev. 2014 Oct;25(5):551-61. doi: 10.1016/j.cytogfr.2014.04.008. Epub 2014 May 9. Cytokine Growth Factor Rev. 2014. PMID: 24861946 Review.
-
Role and fate of PML nuclear bodies in response to interferon and viral infections.Oncogene. 2001 Oct 29;20(49):7274-86. doi: 10.1038/sj.onc.1204854. Oncogene. 2001. PMID: 11704856 Review.
Cited by
-
Incoming human papillomavirus 16 genome is lost in PML protein-deficient HaCaT keratinocytes.Cell Microbiol. 2017 May;19(5):10.1111/cmi.12708. doi: 10.1111/cmi.12708. Epub 2017 Jan 23. Cell Microbiol. 2017. PMID: 27860076 Free PMC article.
-
The prolyl isomerase Pin1 regulates the NF-kappaB signaling pathway and interleukin-8 expression in glioblastoma.Oncogene. 2009 Oct 22;28(42):3735-45. doi: 10.1038/onc.2009.232. Epub 2009 Aug 10. Oncogene. 2009. PMID: 19668231 Free PMC article.
-
Interaction of promyelocytic leukemia/p53 affects signal transducer and activator of transcription-3 activity in response to oncostatin M.Korean J Physiol Pharmacol. 2020 May 1;24(3):203-212. doi: 10.4196/kjpp.2020.24.3.203. Korean J Physiol Pharmacol. 2020. PMID: 32392911 Free PMC article.
-
PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool.Proc Natl Acad Sci U S A. 2009 Mar 24;106(12):4725-30. doi: 10.1073/pnas.0807640106. Epub 2009 Mar 4. Proc Natl Acad Sci U S A. 2009. PMID: 19261859 Free PMC article.
-
Promyelocytic Leukemia Protein Isoform II Promotes Transcription Factor Recruitment To Activate Interferon Beta and Interferon-Responsive Gene Expression.Mol Cell Biol. 2015 May;35(10):1660-72. doi: 10.1128/MCB.01478-14. Epub 2015 Mar 2. Mol Cell Biol. 2015. PMID: 25733689 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

