The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis
- PMID: 17085683
- PMCID: PMC1693942
- DOI: 10.1105/tpc.106.046508
The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis
Abstract
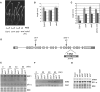
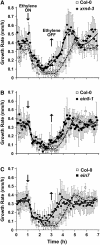
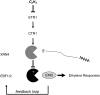
EXORIBONUCLEASE4 (XRN4), the Arabidopsis thaliana homolog of yeast XRN1, is involved in the degradation of several unstable mRNAs. Although a role for XRN4 in RNA silencing of certain transgenes has been reported, xrn4 mutant plants were found to lack any apparent visible phenotype. Here, we show that XRN4 is allelic to the unidentified components of the ethylene response pathway ETHYLENE-INSENSITIVE5/ACC-INSENSITIVE1 (EIN5/AIN1) and EIN7. xrn4 mutant seedlings are ethylene-insensitive as a consequence of the upregulation of EIN3 BINDING F-BOX PROTEIN1 (EBF1) and EBF2 mRNA levels, which encode related F-box proteins involved in the turnover of EIN3 protein, a crucial transcriptional regulator of the ethylene response pathway. Epistasis analysis placed XRN4/EIN5/AIN1 downstream of CTR1 and upstream of EBF1/2. XRN4 does not appear to regulate ethylene signaling via an RNA-INDUCED SILENCING COMPLEX-based RNA silencing mechanism but acts by independent means. The identification of XRN4 as an integral new component in ethylene signaling adds RNA degradation as another posttranscriptional process that modulates the perception of this plant hormone.
Figures


Similar articles
-
ETHYLENE-INSENSITIVE5 encodes a 5'-->3' exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2.Proc Natl Acad Sci U S A. 2006 Sep 5;103(36):13286-93. doi: 10.1073/pnas.0605528103. Epub 2006 Aug 18. Proc Natl Acad Sci U S A. 2006. PMID: 16920797 Free PMC article.
-
The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling.Plant Cell. 2007 Feb;19(2):509-23. doi: 10.1105/tpc.106.048140. Epub 2007 Feb 16. Plant Cell. 2007. PMID: 17307926 Free PMC article.
-
Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories.RNA. 2011 Mar;17(3):501-11. doi: 10.1261/rna.2467911. Epub 2011 Jan 11. RNA. 2011. PMID: 21224377 Free PMC article.
-
Ethylene signaling: new levels of complexity and regulation.Curr Opin Plant Biol. 2008 Oct;11(5):479-85. doi: 10.1016/j.pbi.2008.06.011. Epub 2008 Aug 7. Curr Opin Plant Biol. 2008. PMID: 18692429 Free PMC article. Review.
-
Seedlings Transduce the Depth and Mechanical Pressure of Covering Soil Using COP1 and Ethylene to Regulate EBF1/EBF2 for Soil Emergence.Curr Biol. 2016 Jan 25;26(2):139-149. doi: 10.1016/j.cub.2015.11.053. Epub 2015 Dec 31. Curr Biol. 2016. PMID: 26748855 Free PMC article. Review.
Cited by
-
XRN 5'→3' exoribonucleases: structure, mechanisms and functions.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):590-603. doi: 10.1016/j.bbagrm.2013.03.005. Epub 2013 Mar 19. Biochim Biophys Acta. 2013. PMID: 23517755 Free PMC article. Review.
-
Endoribonuclease DNE1 Promotes Ethylene Response by Modulating EBF1/2 mRNA Processing in Arabidopsis.Int J Mol Sci. 2024 Feb 10;25(4):2138. doi: 10.3390/ijms25042138. Int J Mol Sci. 2024. PMID: 38396815 Free PMC article.
-
Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling.PLoS Genet. 2012;8(11):e1002997. doi: 10.1371/journal.pgen.1002997. Epub 2012 Nov 15. PLoS Genet. 2012. PMID: 23166504 Free PMC article.
-
The 5'-3' Exoribonuclease XRN4 Regulates Auxin Response via the Degradation of Auxin Receptor Transcripts.Genes (Basel). 2018 Dec 17;9(12):638. doi: 10.3390/genes9120638. Genes (Basel). 2018. PMID: 30563022 Free PMC article.
-
The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels.Plant J. 2009 Jan;57(2):332-45. doi: 10.1111/j.1365-313X.2008.03693.x. Epub 2008 Oct 30. Plant J. 2009. PMID: 18808454 Free PMC article.
References
-
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152. - PubMed
-
- Alonso, J.M., and Stepanova, A.N. (2004). The ethylene signaling pathway. Science 306 1513–1515. - PubMed
-
- Boutet, S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J.B., Crete, P., Chen, X., and Vaucheret, H. (2003). Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13 843–848. - PMC - PubMed
NOTE ADDED IN PROOF
-
- While this manuscript was under review, a report by Olmeda et al. (2006) identified EIN5 as XRN4.
-
- Olmedo, G., Guo, H., Gregory, B.D., Nourizadeh, S.D., Aguilar-Henonin, L., Li, H., An, F., Guzman, P., and Ecker, J.R. (2006). ETHYLENE INSENSITIVE5 encodes a 5′ →3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. USA 103 13286–13293. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

