Carotenoid biosynthesis in the primitive red alga Cyanidioschyzon merolae
- PMID: 17085635
- PMCID: PMC1828917
- DOI: 10.1128/EC.00265-06
Carotenoid biosynthesis in the primitive red alga Cyanidioschyzon merolae
Abstract
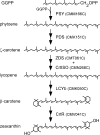
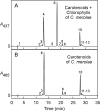
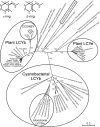
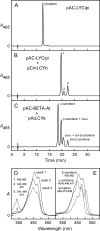
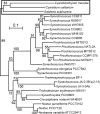
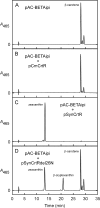
Cyanidioschyzon merolae is considered to be one of the most primitive of eukaryotic photosynthetic organisms. To obtain insights into the origin and evolution of the pathway of carotenoid biosynthesis in eukaryotic plants, the carotenoid content of C. merolae was ascertained, genes encoding enzymes of carotenoid biosynthesis in this unicellular red alga were identified, and the activities of two candidate pathway enzymes of particular interest, lycopene cyclase and beta-carotene hydroxylase, were examined. C. merolae contains perhaps the simplest assortment of chlorophylls and carotenoids found in any eukaryotic photosynthetic organism: chlorophyll a, beta-carotene, and zeaxanthin. Carotenoids with epsilon-rings (e.g., lutein), found in many other red algae and in green algae and land plants, were not detected, and the lycopene cyclase of C. merolae quite specifically produced only beta-ringed carotenoids when provided with lycopene as the substrate in Escherichia coli. Lycopene beta-ring cyclases from several bacteria, cyanobacteria, and land plants also proved to be high-fidelity enzymes, whereas the structurally related epsilon-ring cyclases from several plant species were found to be less specific, yielding products with beta-rings as well as epsilon-rings. C. merolae lacks orthologs of genes that encode the two types of beta-carotene hydroxylase found in land plants, one a nonheme diiron oxygenase and the other a cytochrome P450. A C. merolae chloroplast gene specifies a polypeptide similar to members of a third class of beta-carotene hydroxylases, common in cyanobacteria, but this gene did not produce an active enzyme when expressed in E. coli. The identity of the C. merolae beta-carotene hydroxylase therefore remains uncertain.
Figures


Similar articles
-
Functional Lycopene Cyclase (CruA) in Cyanobacterium, Arthrospira platensis NIES-39, and its Role in Carotenoid Synthesis.Plant Cell Physiol. 2017 Apr 1;58(4):831-838. doi: 10.1093/pcp/pcx015. Plant Cell Physiol. 2017. PMID: 28371918
-
Cloning and Functional Characterization of a Lycopene β-Cyclase from Macrophytic Red Alga Bangia fuscopurpurea.Mar Drugs. 2017 Apr 11;15(4):116. doi: 10.3390/md15040116. Mar Drugs. 2017. PMID: 28398223 Free PMC article.
-
Carotenoid analysis of a liverwort Marchantia polymorpha and functional identification of its lycopene β- and ε-cyclase genes.Plant Cell Physiol. 2014 Jan;55(1):194-200. doi: 10.1093/pcp/pct170. Epub 2013 Nov 26. Plant Cell Physiol. 2014. PMID: 24285752
-
Progress in understanding the origin and functions of carotenoid hydroxylases in plants.Arch Biochem Biophys. 2004 Oct 1;430(1):22-9. doi: 10.1016/j.abb.2004.02.003. Arch Biochem Biophys. 2004. PMID: 15325908 Review.
-
Genetic manipulation of carotenoid biosynthesis and photoprotection.Philos Trans R Soc Lond B Biol Sci. 2000 Oct 29;355(1402):1395-403. doi: 10.1098/rstb.2000.0701. Philos Trans R Soc Lond B Biol Sci. 2000. PMID: 11127994 Free PMC article. Review.
Cited by
-
Inactivation of genes encoding plastoglobuli-like proteins in Synechocystis sp. PCC 6803 leads to a light-sensitive phenotype.J Bacteriol. 2010 Mar;192(6):1700-9. doi: 10.1128/JB.01434-09. Epub 2010 Jan 15. J Bacteriol. 2010. PMID: 20081034 Free PMC article.
-
The carbon-concentrating mechanism of the extremophilic red microalga Cyanidioschyzon merolae.Photosynth Res. 2023 May;156(2):247-264. doi: 10.1007/s11120-023-01000-6. Epub 2023 Feb 13. Photosynth Res. 2023. PMID: 36780115 Free PMC article.
-
Two-step pathway for isoprenoid synthesis.Proc Natl Acad Sci U S A. 2019 Jan 8;116(2):506-511. doi: 10.1073/pnas.1812935116. Epub 2018 Dec 24. Proc Natl Acad Sci U S A. 2019. PMID: 30584096 Free PMC article.
-
Carotenoids Database: structures, chemical fingerprints and distribution among organisms.Database (Oxford). 2017 Jan 1;2017(1):bax004. doi: 10.1093/database/bax004. Database (Oxford). 2017. PMID: 28365725 Free PMC article.
-
Exogenous Arachidonic Acid Affects Fucoxanthin Biosynthesis and Photoprotection in Phaeodactylum tricornutum.Mar Drugs. 2022 Oct 17;20(10):644. doi: 10.3390/md20100644. Mar Drugs. 2022. PMID: 36286467 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Antoine, R., and C. Locht. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6:1785-1799. - PubMed
-
- Barbier, G., C. Oesterhelt, M. D. Larson, R. G. Halgren, C. Wilkerson, R. M. Garavito, C. Benning, and A. P. M. Weber. 2005. Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 137:460-474. - PMC - PubMed
-
- Bjørnland, T., and M. Aguilar-Martinez. 1976. Carotenoids in red algae. Phytochemistry 15:291-296.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

