U12DB: a database of orthologous U12-type spliceosomal introns
- PMID: 17082203
- PMCID: PMC1635337
- DOI: 10.1093/nar/gkl796
U12DB: a database of orthologous U12-type spliceosomal introns
Abstract
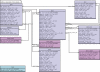
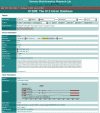
U12-type introns are spliced by the U12-dependent spliceosome and are present in the genomes of many higher eukaryotic lineages including plants, chordates and some invertebrates. However, due to their relatively recent discovery and a systematic bias against recognition of non-canonical splice sites in general, the introns defined by U12-type splice sites are under-represented in genome annotations. Such under-representation compounds the already difficult problem of determining gene structures. It also impedes attempts to study these introns genome-wide or phylum-wide. The resource described here, the U12 Intron Database (U12DB), aims to catalog the U12-type introns of completely sequenced eukaryotic genomes in a framework that groups orthologous introns with each other. This will aid further investigations into the evolution and mechanism of U12-dependent splicing as well as assist ongoing genome annotation efforts. Public access to the U12DB is available at http://genome.imim.es/cgi-bin/u12db/u12db.cgi.
Figures


Similar articles
-
Splicing of a rare class of introns by the U12-dependent spliceosome.Biol Chem. 2005 Aug;386(8):713-24. doi: 10.1515/BC.2005.084. Biol Chem. 2005. PMID: 16201866 Review.
-
RS domain-splicing signal interactions in splicing of U12-type and U2-type introns.Nat Struct Mol Biol. 2007 Jul;14(7):597-603. doi: 10.1038/nsmb1263. Epub 2007 Jul 1. Nat Struct Mol Biol. 2007. PMID: 17603499
-
Identification of genome-wide non-canonical spliced regions and analysis of biological functions for spliced sequences using Read-Split-Fly.BMC Bioinformatics. 2017 Oct 3;18(Suppl 11):382. doi: 10.1186/s12859-017-1801-y. BMC Bioinformatics. 2017. PMID: 28984182 Free PMC article.
-
Alternative splicing and bioinformatic analysis of human U12-type introns.Nucleic Acids Res. 2007;35(6):1833-41. doi: 10.1093/nar/gkm026. Epub 2007 Mar 1. Nucleic Acids Res. 2007. PMID: 17332017 Free PMC article.
-
Coevolution of genomic intron number and splice sites.Trends Genet. 2007 Jul;23(7):321-5. doi: 10.1016/j.tig.2007.04.001. Epub 2007 Apr 18. Trends Genet. 2007. PMID: 17442445 Review.
Cited by
-
Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome.Nat Commun. 2015 Jan 14;6:6042. doi: 10.1038/ncomms7042. Nat Commun. 2015. PMID: 25586593 Free PMC article.
-
The minor spliceosomal protein U11/U12-31K is an RNA chaperone crucial for U12 intron splicing and the development of dicot and monocot plants.PLoS One. 2012;7(8):e43707. doi: 10.1371/journal.pone.0043707. Epub 2012 Aug 17. PLoS One. 2012. PMID: 22912901 Free PMC article.
-
Minor intron splicing revisited: identification of new minor intron-containing genes and tissue-dependent retention and alternative splicing of minor introns.BMC Genomics. 2019 Aug 30;20(1):686. doi: 10.1186/s12864-019-6046-x. BMC Genomics. 2019. PMID: 31470809 Free PMC article.
-
Dysregulated minor intron splicing in cancer.Cancer Sci. 2022 Sep;113(9):2934-2942. doi: 10.1111/cas.15476. Epub 2022 Jul 11. Cancer Sci. 2022. PMID: 35766428 Free PMC article. Review.
-
Splicing efficiency of minor introns in a mouse model of SMA predominantly depends on their branchpoint sequence and can involve the contribution of major spliceosome components.RNA. 2022 Mar;28(3):303-319. doi: 10.1261/rna.078329.120. Epub 2021 Dec 10. RNA. 2022. PMID: 34893560 Free PMC article.
References
-
- Patel A.A., Steitz J.A. Splicing double: insights from the second spliceosome. Nature Rev. Mol. Cell Biol. 2003;4:960–970. - PubMed
-
- Will C.L., Luhrmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 2005;386:713–724. - PubMed
-
- Hall S.L., Padgett R.A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol. 1994;239:357–365. - PubMed

