Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity
- PMID: 17081564
- PMCID: PMC1832103
- DOI: 10.1016/j.jmb.2006.09.083
Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity
Abstract
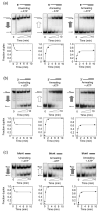
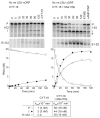
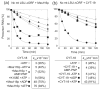
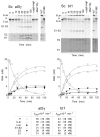
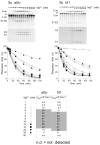
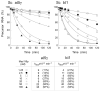
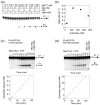
The RNA-catalyzed splicing of group I and group II introns is facilitated by proteins that stabilize the active RNA structure or act as RNA chaperones to disrupt stable inactive structures that are kinetic traps in RNA folding. In Neurospora crassa and Saccharomyces cerevisiae, the latter function is fulfilled by specific DEAD-box proteins, denoted CYT-19 and Mss116p, respectively. Previous studies showed that purified CYT-19 stimulates the in vitro splicing of structurally diverse group I and group II introns, and uses the energy of ATP binding or hydrolysis to resolve kinetic traps. Here, we purified Mss116p and show that it has RNA-dependent ATPase activity, unwinds RNA duplexes in a non-polar fashion, and promotes ATP-independent strand-annealing. Further, we show that Mss116p binds RNA non-specifically and promotes in vitro splicing of both group I and group II intron RNAs, as well as RNA cleavage by the aI5gamma-derived D135 ribozyme. However, Mss116p also has ATP hydrolysis-independent effects on some of these reactions, which are not shared by CYT-19 and may reflect differences in its RNA-binding properties. We also show that a non-mitochondrial DEAD-box protein, yeast Ded1p, can function almost as efficiently as CYT-19 and Mss116p in splicing the yeast aI5gamma group II intron and less efficiently in splicing the bI1 group II intron. Together, our results show that Mss116p, like CYT-19, can act broadly as an RNA chaperone to stimulate the splicing of diverse group I and group II introns, and that Ded1p also has an RNA chaperone activity that can be assayed by its effect on splicing mitochondrial introns. Nevertheless, these DEAD-box protein RNA chaperones are not completely interchangeable and appear to function in somewhat different ways, using biochemical activities that have likely been tuned by coevolution to function optimally on specific RNA substrates.
Figures

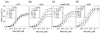


Similar articles
-
ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo.J Mol Biol. 2011 Aug 19;411(3):661-79. doi: 10.1016/j.jmb.2011.05.047. Epub 2011 Jun 7. J Mol Biol. 2011. PMID: 21679717 Free PMC article.
-
Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones.J Mol Biol. 2009 Jun 19;389(4):674-93. doi: 10.1016/j.jmb.2009.04.043. Epub 2009 Apr 23. J Mol Biol. 2009. PMID: 19393667 Free PMC article.
-
Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro.J Mol Biol. 2008 Feb 1;375(5):1344-64. doi: 10.1016/j.jmb.2007.11.041. Epub 2007 Nov 22. J Mol Biol. 2008. PMID: 18096186 Free PMC article.
-
Mss116p: a DEAD-box protein facilitates RNA folding.RNA Biol. 2013 Jan;10(1):71-82. doi: 10.4161/rna.22492. Epub 2012 Oct 12. RNA Biol. 2013. PMID: 23064153 Free PMC article. Review.
-
DEAD-box proteins as RNA helicases and chaperones.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):135-52. doi: 10.1002/wrna.50. Wiley Interdiscip Rev RNA. 2011. PMID: 21297876 Free PMC article. Review.
Cited by
-
The Thermus thermophilus DEAD box helicase Hera contains a modified RNA recognition motif domain loosely connected to the helicase core.RNA. 2009 Nov;15(11):1993-2001. doi: 10.1261/rna.1820009. Epub 2009 Aug 26. RNA. 2009. PMID: 19710183 Free PMC article.
-
Evaluating target silencing by short hairpin RNA mediated by the group I intron in cultured mammalian cells.BMC Biotechnol. 2011 Jul 25;11:79. doi: 10.1186/1472-6750-11-79. BMC Biotechnol. 2011. PMID: 21781346 Free PMC article.
-
A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins.Mol Cell Biol. 2008 May;28(10):3359-71. doi: 10.1128/MCB.01555-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332124 Free PMC article.
-
The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly.J Mol Biol. 2013 Oct 23;425(20):3824-38. doi: 10.1016/j.jmb.2013.05.016. Epub 2013 May 28. J Mol Biol. 2013. PMID: 23721653 Free PMC article.
-
RNA catalysis as a probe for chaperone activity of DEAD-box helicases.Methods Enzymol. 2012;511:111-30. doi: 10.1016/B978-0-12-396546-2.00005-X. Methods Enzymol. 2012. PMID: 22713317 Free PMC article.
References
-
- Lambowitz AM, Caprara MG, Zimmerly S, Perlman PS. Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2. Cold Spring Harbor Laboratory Press: Plainview, NY; 1999. pp. 451–485.
-
- Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol. 2003;38:249–303. - PubMed
-
- Pyle AM, Lambowitz AM. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3. Cold Spring Harbor Laboratory Press; Plainview, NY: 2006. pp. 469–505.
-
- Caprara MG, Nilsen TW. RNA: versatility in form and function. Nat Struct Biol. 2000;7:831–833. - PubMed
-
- Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

