The requirement of sterol glucoside for pexophagy in yeast is dependent on the species and nature of peroxisome inducers
- PMID: 17079731
- PMCID: PMC1751328
- DOI: 10.1091/mbc.e06-06-0554
The requirement of sterol glucoside for pexophagy in yeast is dependent on the species and nature of peroxisome inducers
Abstract
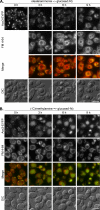
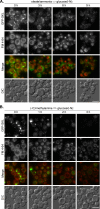
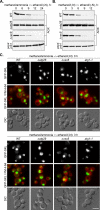
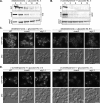
Sterol glucosyltransferase, Ugt51/Atg26, is essential for both micropexophagy and macropexophagy of methanol-induced peroxisomes in Pichia pastoris. However, the role of this protein in pexophagy in other yeast remained unclear. We show that oleate- and amine-induced peroxisomes in Yarrowia lipolytica are degraded by Atg26-independent macropexophagy. Surprisingly, Atg26 was also not essential for macropexophagy of oleate- and amine-induced peroxisomes in P. pastoris, suggesting that the function of sterol glucoside (SG) in pexophagy is both species and peroxisome inducer specific. However, the rates of degradation of oleate- and amine-induced peroxisomes in P. pastoris were reduced in the absence of SG, indicating that P. pastoris specifically uses sterol conversion by Atg26 to enhance selective degradation of peroxisomes. However, methanol-induced peroxisomes apparently have lost the redundant ability to be degraded without SG. We also show that the P. pastoris Vac8 armadillo repeat protein is not essential for macropexophagy of methanol-, oleate-, or amine-induced peroxisomes, which makes PpVac8 the first known protein required for the micropexophagy, but not for the macropexophagy, machinery. The uniqueness of Atg26 and Vac8 functions under different pexophagy conditions demonstrates that not only pexophagy inducers, such as glucose or ethanol, but also the inducers of peroxisomes, such as methanol, oleate, or primary amines, determine the requirements for subsequent pexophagy in yeast.
Figures




Similar articles
-
Autophagy-related pathways and specific role of sterol glucoside in yeasts.Autophagy. 2007 May-Jun;3(3):263-5. doi: 10.4161/auto.3907. Epub 2007 May 23. Autophagy. 2007. PMID: 17329963
-
Sterol glucosyltransferases have different functional roles in Pichia pastoris and Yarrowia lipolytica.Cell Biol Int. 2003;27(11):947-52. doi: 10.1016/j.cellbi.2003.08.004. Cell Biol Int. 2003. PMID: 14585290
-
Atg28, a novel coiled-coil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris.Autophagy. 2006 Jan-Mar;2(1):30-8. doi: 10.4161/auto.2226. Epub 2006 Jan 9. Autophagy. 2006. PMID: 16874081
-
Pexophagy: the selective autophagy of peroxisomes.Autophagy. 2005 Jul;1(2):75-83. doi: 10.4161/auto.1.2.1737. Epub 2005 Jul 13. Autophagy. 2005. PMID: 16874024 Review.
-
Pexophagy: autophagic degradation of peroxisomes.Biochim Biophys Acta. 2006 Dec;1763(12):1767-75. doi: 10.1016/j.bbamcr.2006.08.023. Epub 2006 Aug 24. Biochim Biophys Acta. 2006. PMID: 17005271 Review.
Cited by
-
Pexophagy: the selective degradation of peroxisomes.Int J Cell Biol. 2012;2012:512721. doi: 10.1155/2012/512721. Epub 2012 Mar 27. Int J Cell Biol. 2012. PMID: 22536249 Free PMC article.
-
Autophagic processes in yeast: mechanism, machinery and regulation.Genetics. 2013 Jun;194(2):341-61. doi: 10.1534/genetics.112.149013. Genetics. 2013. PMID: 23733851 Free PMC article. Review.
-
The emerging role of autophagy in peroxisome dynamics and lipid metabolism of phyllosphere microorganisms.Front Plant Sci. 2014 Mar 11;5:81. doi: 10.3389/fpls.2014.00081. eCollection 2014. Front Plant Sci. 2014. PMID: 24653730 Free PMC article. Review.
-
Redox regulated peroxisome homeostasis.Redox Biol. 2015;4:104-8. doi: 10.1016/j.redox.2014.12.006. Epub 2014 Dec 18. Redox Biol. 2015. PMID: 25545794 Free PMC article. Review.
-
Catabolite repression of Aox in Pichia pastoris is dependent on hexose transporter PpHxt1 and pexophagy.Appl Environ Microbiol. 2010 Sep;76(18):6108-18. doi: 10.1128/AEM.00607-10. Epub 2010 Jul 23. Appl Environ Microbiol. 2010. PMID: 20656869 Free PMC article.
References
-
- Ano Y., Hattori T., Kato N., Sakai Y. Intracellular ATP correlates with mode of pexophagy in Pichia pastoris. Biosci. Biotechnol. Biochem. 2005;69:1527–1533. - PubMed
-
- Baerends R. J., Faber K. N., Kram A. M., Kiel J. A., van der Klei I. J., Veenhuis M. A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J. Biol. Chem. 2000;275:9986–9995. - PubMed
-
- Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Nonconventional Yeasts in Biotechnology. Berlin, Germany: Springer; 1996. pp. 313–388.
-
- Bellu A. R., Kiel J. A. Selective degradation of peroxisomes in yeasts. Microsc. Res. Tech. 2003;61:161–170. - PubMed

