Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes
- PMID: 17041050
- PMCID: PMC1797373
- DOI: 10.1128/JB.00976-06
Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes
Abstract
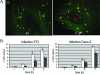
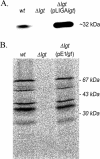
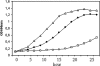
Lipoprotein anchoring in bacteria is mediated by the prolipoprotein diacylglyceryl transferase (Lgt), which catalyzes the transfer of a diacylglyceryl moiety to the prospective N-terminal cysteine of the mature lipoprotein. Deletion of the lgt gene in the gram-positive pathogen Listeria monocytogenes (i) impairs intracellular growth of the bacterium in different eukaryotic cell lines and (ii) leads to increased release of lipoproteins into the culture supernatant. Comparative extracellular proteome analyses of the EGDe wild-type strain and the Delta lgt mutant provided systematic insight into the relative expression of lipoproteins. Twenty-six of the 68 predicted lipoproteins were specifically released into the extracellular proteome of the Delta lgt strain, and this proved that deletion of lgt is an excellent approach for experimental verification of listerial lipoproteins. Consequently, we generated Delta lgt Delta prfA double mutants to detect lipoproteins belonging to the main virulence regulon that is controlled by PrfA. Overall, we identified three lipoproteins whose extracellular levels are regulated and one lipoprotein that is posttranslationally modified depending on PrfA. It is noteworthy that in contrast to previous studies of Escherichia coli, we unambiguously demonstrated that lipidation by Lgt is not a prerequisite for activity of the lipoprotein-specific signal peptidase II (Lsp) in Listeria.
Figures
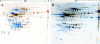


Comment in
-
An important step in listeria lipoprotein research.J Bacteriol. 2007 Jan;189(2):294-7. doi: 10.1128/JB.01577-06. Epub 2006 Oct 27. J Bacteriol. 2007. PMID: 17071755 Free PMC article. No abstract available.
Similar articles
-
Role of prolipoprotein diacylglyceryl transferase (Lgt) and lipoprotein-specific signal peptidase II (LspA) in localization and physiological function of lipoprotein MsmE in Streptococcus mutans.Oral Microbiol Immunol. 2008 Dec;23(6):515-9. doi: 10.1111/j.1399-302X.2008.00455.x. Oral Microbiol Immunol. 2008. PMID: 18954360
-
New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA.Mol Microbiol. 2004 Jun;52(6):1553-65. doi: 10.1111/j.1365-2958.2004.04052.x. Mol Microbiol. 2004. PMID: 15186408
-
Negative control of Listeria monocytogenes virulence genes by a diffusible autorepressor.Mol Microbiol. 2004 Apr;52(2):601-11. doi: 10.1111/j.1365-2958.2004.04003.x. Mol Microbiol. 2004. PMID: 15066044
-
Regulation of virulence genes in Listeria.Int J Med Microbiol. 2001 May;291(2):145-57. doi: 10.1078/1438-4221-00111. Int J Med Microbiol. 2001. PMID: 11437337 Review.
-
The PrfA virulence regulon.Microbes Infect. 2007 Aug;9(10):1196-207. doi: 10.1016/j.micinf.2007.05.007. Epub 2007 May 7. Microbes Infect. 2007. PMID: 17764998 Review.
Cited by
-
Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway.J Bacteriol. 2007 Jul;189(14):5090-100. doi: 10.1128/JB.00163-07. Epub 2007 May 11. J Bacteriol. 2007. PMID: 17496088 Free PMC article.
-
Functional analyses of mycobacterial lipoprotein diacylglyceryl transferase and comparative secretome analysis of a mycobacterial lgt mutant.J Bacteriol. 2012 Aug;194(15):3938-49. doi: 10.1128/JB.00127-12. Epub 2012 May 18. J Bacteriol. 2012. PMID: 22609911 Free PMC article.
-
The iron-regulated staphylococcal lipoproteins.Front Cell Infect Microbiol. 2012 Apr 4;2:41. doi: 10.3389/fcimb.2012.00041. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919632 Free PMC article. Review.
-
N-Terminal clustering of the O-glycosylation sites in the Mycobacterium tuberculosis lipoprotein SodC.Glycobiology. 2009 Jan;19(1):38-51. doi: 10.1093/glycob/cwn102. Epub 2008 Oct 8. Glycobiology. 2009. PMID: 18842962 Free PMC article.
-
A structural comparison of Listeria monocytogenes protein chaperones PrsA1 and PrsA2 reveals molecular features required for virulence.Mol Microbiol. 2016 Jul;101(1):42-61. doi: 10.1111/mmi.13367. Epub 2016 May 27. Mol Microbiol. 2016. PMID: 27007641 Free PMC article.
References
-
- Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science 285:736-739. - PubMed
-
- Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. - PubMed
-
- Arai, M., M. Ikeda, and T. Shimizu. 2003. Comprehensive analysis of transmembrane topologies in prokaryotic genomes. Gene 304:77-86. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantiation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

