Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases
- PMID: 17041027
- PMCID: PMC1676070
- DOI: 10.1104/pp.106.088476
Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases
Abstract
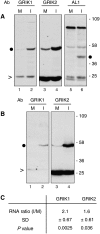
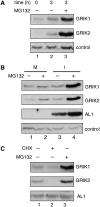
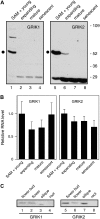
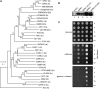
Geminivirus Rep-interacting kinase 1 (GRIK1) and GRIK2 constitute a small protein kinase family in Arabidopsis (Arabidopsis thaliana). An earlier study showed that a truncated version of GRIK1 binds to the geminivirus replication protein AL1. We show here both full-length GRIK1 and GRIK2 interact with AL1 in yeast two-hybrid studies. Using specific antibodies, we showed that both Arabidopsis kinases are elevated in infected leaves. Immunoblot analysis of healthy plants revealed that GRIK1 and GRIK2 are highest in young leaf and floral tissues and low or undetectable in mature tissues. Immunohistochemical staining showed that the kinases accumulate in the shoot apical meristem, leaf primordium, and emerging petiole. Unlike the protein patterns, GRIK1 and GRIK2 transcript levels only show a small increase during infection and do not change significantly during development. Treating healthy seedlings and infected leaves with the proteasome inhibitor MG132 resulted in higher GRIK1 and GRIK2 protein levels, whereas treatment with the translation inhibitor cycloheximide reduced both kinases, demonstrating that their accumulation is modulated by posttranscriptional processes. Phylogenetic comparisons indicated that GRIK1, GRIK2, and related kinases from Medicago truncatula and rice (Oryza sativa) are most similar to the yeast kinases PAK1, TOS3, and ELM1 and the mammalian kinase CaMKK, which activate the yeast kinase SNF1 and its mammalian homolog AMPK, respectively. Complementation studies using a PAK1/TOS3/ELM1 triple mutant showed that GRIK1 and GRIK2 can functionally replace the yeast kinases, suggesting that the Arabidopsis kinases mediate one or more processes during early plant development and geminivirus infection by activating SNF1-related kinases.
Figures




Similar articles
-
Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop.Plant Physiol. 2009 Jun;150(2):996-1005. doi: 10.1104/pp.108.132787. Epub 2009 Apr 1. Plant Physiol. 2009. PMID: 19339507 Free PMC article.
-
Upstream kinases of plant SnRKs are involved in salt stress tolerance.Plant J. 2018 Jan;93(1):107-118. doi: 10.1111/tpj.13761. Epub 2017 Dec 2. Plant J. 2018. PMID: 29094495 Free PMC article.
-
A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection.Plant Cell. 2002 Aug;14(8):1817-32. doi: 10.1105/tpc.003681. Plant Cell. 2002. PMID: 12172024 Free PMC article.
-
Plant SnRK1 Kinases: Structure, Regulation, and Function.Exp Suppl. 2016;107:403-438. doi: 10.1007/978-3-319-43589-3_17. Exp Suppl. 2016. PMID: 27812990 Review.
-
Molecular Insights into the Enigmatic Metabolic Regulator, SnRK1.Trends Plant Sci. 2016 Apr;21(4):341-353. doi: 10.1016/j.tplants.2015.11.001. Epub 2015 Nov 28. Trends Plant Sci. 2016. PMID: 26642889 Review.
Cited by
-
Subtle Regulation of Potato Acid Invertase Activity by a Protein Complex of Invertase, Invertase Inhibitor, and SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE.Plant Physiol. 2015 Aug;168(4):1807-19. doi: 10.1104/pp.15.00664. Epub 2015 Jul 1. Plant Physiol. 2015. PMID: 26134163 Free PMC article.
-
DNA-binding protein phosphatase AtDBP1 mediates susceptibility to two potyviruses in Arabidopsis.Plant Physiol. 2010 Aug;153(4):1521-5. doi: 10.1104/pp.110.158923. Epub 2010 May 27. Plant Physiol. 2010. PMID: 20508138 Free PMC article. No abstract available.
-
Functional analysis of a novel motif conserved across geminivirus Rep proteins.J Virol. 2011 Feb;85(3):1182-92. doi: 10.1128/JVI.02143-10. Epub 2010 Nov 17. J Virol. 2011. PMID: 21084480 Free PMC article.
-
A New Type of Satellite Associated with Cassava Mosaic Begomoviruses.J Virol. 2021 Oct 13;95(21):e0043221. doi: 10.1128/JVI.00432-21. Epub 2021 Aug 18. J Virol. 2021. PMID: 34406866 Free PMC article.
-
Rice stripe tenuivirus nonstructural protein 3 hijacks the 26S proteasome of the small brown planthopper via direct interaction with regulatory particle non-ATPase subunit 3.J Virol. 2015 Apr;89(8):4296-310. doi: 10.1128/JVI.03055-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653432 Free PMC article.
References
-
- Bisaro DM (2006) Silencing suppression by geminivirus proteins. Virology 344: 158–168 - PubMed
-
- Castillo AG, Collinet D, Deret S, Kashoggi A, Bejarano ER (2003) Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312: 381–394 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

