Determination of thermodynamics and kinetics of RNA reactions by force
- PMID: 17040613
- PMCID: PMC2542947
- DOI: 10.1017/S0033583506004446
Determination of thermodynamics and kinetics of RNA reactions by force
Abstract
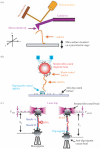
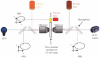
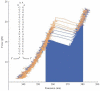
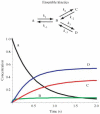
Single-molecule methods have made it possible to apply force to an individual RNA molecule. Two beads are attached to the RNA; one is on a micropipette, the other is in a laser trap. The force on the RNA and the distance between the beads are measured. Force can change the equilibrium and the rate of any reaction in which the product has a different extension from the reactant. This review describes use of laser tweezers to measure thermodynamics and kinetics of unfolding/refolding RNA. For a reversible reaction the work directly provides the free energy; for irreversible reactions the free energy is obtained from the distribution of work values. The rate constants for the folding and unfolding reactions can be measured by several methods. The effect of pulling rate on the distribution of force-unfolding values leads to rate constants for unfolding. Hopping of the RNA between folded and unfolded states at constant force provides both unfolding and folding rates. Force-jumps and force-drops, similar to the temperature jump method, provide direct measurement of reaction rates over a wide range of forces. The advantages of applying force and using single-molecule methods are discussed. These methods, for example, allow reactions to be studied in non-denaturing solvents at physiological temperatures; they also simplify analysis of kinetic mechanisms because only one intermediate at a time is present. Unfolding of RNA in biological cells by helicases, or ribosomes, has similarities to unfolding by force.
Figures







References
-
- Allison D, Hinterdorfer P, Han W. Biomolecular force measurements and the atomic force microscope. Current Opinion in Biotechnology. 2002;13:47–51. - PubMed
-
- Ashkin A. Methods in Cell Biology. Vol. 55. Academic Press; San Diego: 1998. Forces of a single-beam gradient laser trap on a dielectric sphere in the ray optics regime; pp. 1–27. - PubMed
-
- Ashkin A, Dziedzic J, Bjorkholm J, Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Optical Letters. 1986;11:288–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

