N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination
- PMID: 17034622
- PMCID: PMC1617110
- DOI: 10.1186/1471-2199-7-34
N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination
Abstract
Background: Termination of translation in eukaryotes requires two release factors, eRF1, which recognizes all three nonsense codons and facilitates release of the nascent polypeptide chain, and eRF3 stimulating translation termination in a GTP-depended manner. eRF3 from different organisms possess a highly conservative C region (eRF3C), which is responsible for the function in translation termination, and almost always contain the N-terminal extension, which is inessential and vary both in structure and length. In the yeast Saccharomyces cerevisiae the N-terminal region of eRF3 is responsible for conversion of this protein into the aggregated and functionally inactive prion form.
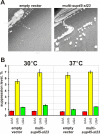
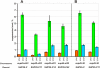
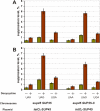
Results: Here, we examined functional importance of the N-terminal region of a non-prion form of yeast eRF3. The screen for mutations which are lethal in combination with the SUP35-C allele encoding eRF3C revealed the sup45 mutations which alter the N-terminal domain of eRF1 and increase nonsense codon readthrough. However, further analysis showed that synthetic lethality was not caused by the increased levels of nonsense codon readthrough. Dominant mutations in SUP35-C were obtained and characterized, which remove its synthetic lethality with the identified sup45 mutations, thus indicating that synthetic lethality was not due to a disruption of interaction with proteins that bind to this eRF3 region.
Conclusion: These and other data demonstrate that the N-terminal region of eRF3 is involved both in modulation of the efficiency of translation termination and functioning of the eRF1/eRF3 complex outside of translation termination.
Figures



Similar articles
-
GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination.Mol Cell Biol. 2004 Sep;24(17):7769-78. doi: 10.1128/MCB.24.17.7769-7778.2004. Mol Cell Biol. 2004. PMID: 15314182 Free PMC article.
-
Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3.EMBO J. 1995 Aug 15;14(16):4065-72. doi: 10.1002/j.1460-2075.1995.tb00078.x. EMBO J. 1995. PMID: 7664746 Free PMC article.
-
[Characterization of missense mutations in the SUP45 gene of Saccharomyces cerevisiae encoding translation termination factor eRF1].Genetika. 2004 May;40(5):599-606. Genetika. 2004. PMID: 15272556 Russian.
-
Modulation of efficiency of translation termination in Saccharomyces cerevisiae.Prion. 2014;8(3):247-60. doi: 10.4161/pri.29851. Epub 2014 Nov 1. Prion. 2014. PMID: 25486049 Free PMC article. Review.
-
Translation termination and its regulation in eukaryotes: recent insights provided by studies in yeast.Biochemistry (Mosc). 1999 Dec;64(12):1360-6. Biochemistry (Mosc). 1999. PMID: 10648959 Review.
Cited by
-
Could yeast prion domains originate from polyQ/N tracts?Prion. 2013 May-Jun;7(3):209-14. doi: 10.4161/pri.24628. Prion. 2013. PMID: 23764835 Free PMC article.
-
Sex, prions, and plasmids in yeast.Proc Natl Acad Sci U S A. 2012 Oct 2;109(40):E2683-90. doi: 10.1073/pnas.1213449109. Epub 2012 Sep 4. Proc Natl Acad Sci U S A. 2012. PMID: 22949655 Free PMC article.
-
Prions of fungi: inherited structures and biological roles.Nat Rev Microbiol. 2007 Aug;5(8):611-8. doi: 10.1038/nrmicro1708. Nat Rev Microbiol. 2007. PMID: 17632572 Free PMC article. Review.
-
Suicidal [PSI+] is a lethal yeast prion.Proc Natl Acad Sci U S A. 2011 Mar 29;108(13):5337-41. doi: 10.1073/pnas.1102762108. Epub 2011 Mar 14. Proc Natl Acad Sci U S A. 2011. PMID: 21402947 Free PMC article.
-
Interdependence of amyloid formation in yeast: implications for polyglutamine disorders and biological functions.Prion. 2010 Jan-Mar;4(1):45-52. doi: 10.4161/pri.4.1.11074. Epub 2010 Jan 18. Prion. 2010. PMID: 20118659 Free PMC article.
References
-
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, Justesen J, Kisselev LL. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

