Analysis of a new human parechovirus allows the definition of parechovirus types and the identification of RNA structural domains
- PMID: 17005640
- PMCID: PMC1797470
- DOI: 10.1128/JVI.00584-06
Analysis of a new human parechovirus allows the definition of parechovirus types and the identification of RNA structural domains
Abstract
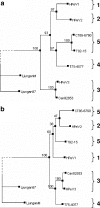
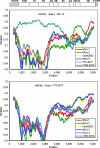
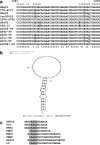
Human parechoviruses (HPeV), members of the Parechovirus genus of Picornaviridae, are frequent pathogens but have been comparatively poorly studied, and little is known of their diversity, evolution, and molecular biology. To increase the amount of information available, we have analyzed 7 HPeV strains isolated in California between 1973 and 1992. We found that, on the basis of VP1 sequences, these fall into two genetic groups, one of which has not been previously observed, bringing the number of known groups to five. While these correlate partly with the three known serotypes, two members of the HPeV2 serotype belong to different genetic groups. In view of the growing importance of molecular techniques in diagnosis, we suggest that genotype is an important criterion for identifying viruses, and we propose that the genetic groups we have defined should be termed human parechovirus types 1 to 5. Complete nucleotide sequence analysis of two of the Californian isolates, representing two types, confirmed the identification of a new genetic group and suggested a role for recombination in parechovirus evolution. It also allowed the identification of a putative HPeV1 cis-acting replication element, which is located in the VP0 coding region, as well as the refinement of previously predicted 5' and 3' untranslated region structures. Thus, the results have significantly improved our understanding of these common pathogens.
Figures


Similar articles
-
Intertypic recombination of human parechovirus 4 isolated from infants with sepsis-like disease.J Clin Virol. 2017 Mar;88:1-7. doi: 10.1016/j.jcv.2017.01.001. Epub 2017 Jan 3. J Clin Virol. 2017. PMID: 28081453
-
Evolution and conservation in human parechovirus genomes.J Gen Virol. 2009 Jul;90(Pt 7):1702-1712. doi: 10.1099/vir.0.008813-0. Epub 2009 Mar 4. J Gen Virol. 2009. PMID: 19264613
-
High prevalence of human Parechovirus (HPeV) genotypes in the Amsterdam region and identification of specific HPeV variants by direct genotyping of stool samples.J Clin Microbiol. 2008 Dec;46(12):3965-70. doi: 10.1128/JCM.01379-08. Epub 2008 Oct 22. J Clin Microbiol. 2008. PMID: 18945833 Free PMC article.
-
Pediatric parechovirus infections.J Clin Virol. 2014 Jun;60(2):84-9. doi: 10.1016/j.jcv.2014.03.003. Epub 2014 Mar 13. J Clin Virol. 2014. PMID: 24690382 Review.
-
Parechovirus A Pathogenesis and the Enigma of Genotype A-3.Viruses. 2019 Nov 14;11(11):1062. doi: 10.3390/v11111062. Viruses. 2019. PMID: 31739613 Free PMC article. Review.
Cited by
-
Genome and infection characteristics of human parechovirus type 1: the interplay between viral infection and type I interferon antiviral system.PLoS One. 2015 Feb 3;10(2):e0116158. doi: 10.1371/journal.pone.0116158. eCollection 2015. PLoS One. 2015. PMID: 25646764 Free PMC article.
-
Initiation of protein-primed picornavirus RNA synthesis.Virus Res. 2015 Aug 3;206:12-26. doi: 10.1016/j.virusres.2014.12.028. Epub 2015 Jan 12. Virus Res. 2015. PMID: 25592245 Free PMC article. Review.
-
A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody.J Virol. 2019 Feb 5;93(4):e01597-18. doi: 10.1128/JVI.01597-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463974 Free PMC article.
-
Human parechovirus infection in children hospitalized with acute gastroenteritis in Sri Lanka.J Clin Microbiol. 2011 Jan;49(1):364-6. doi: 10.1128/JCM.02151-10. Epub 2010 Nov 3. J Clin Microbiol. 2011. PMID: 21048003 Free PMC article.
-
Novel human parechovirus from Brazil.Emerg Infect Dis. 2009 Feb;15(2):310-3. doi: 10.3201/eid1502.081028. Emerg Infect Dis. 2009. PMID: 19193281 Free PMC article.
References
-
- Abed, Y., and G. Boivin. 2005. Molecular characterization of a Canadian human parechovirus (HPeV)-3 isolate and its relationship to other HPeVs. J. Med. Virol. 77:566-570. - PubMed
-
- Benschop, K. S. M., J. Schinkel, R. P. Minnaar, D. Pajkrt, L. Spanjerberg, H. C. Kraakman, B. Berkhout, H. L. Zaaijer, M. G. H. M. Beld, and K. C. Wolthers. 2006. Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin. Infect. Dis. 42:204-210. - PubMed
-
- Chang, K. H., P. Auvinen, T. Hyypiä, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9; implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

