Activation of alpha-tropomyosin exon 2 is regulated by the SR protein 9G8 and heterogeneous nuclear ribonucleoproteins H and F
- PMID: 17000773
- PMCID: PMC1636816
- DOI: 10.1128/MCB.01677-06
Activation of alpha-tropomyosin exon 2 is regulated by the SR protein 9G8 and heterogeneous nuclear ribonucleoproteins H and F
Abstract
The inclusion of exons 2 and 3 of alpha-tropomyosin is governed through tissue-specific alternative splicing. These exons are mutually exclusive, with exon 2 included in smooth muscle cells and exon 3 included in nearly all other cell types. Several cis-acting sequences contribute to this splicing decision: the branchpoints and pyrimidine tracts upstream of both exons, UGC-repeat elements flanking exon 3, and a series of purine-rich enhancers in exon 2. Previous work showed that proteins rich in serine-arginine (SR) dipeptides act through the exon 2 enhancers, but the specific proteins responsible for such activation remained unknown. Here we show that a 35-kDa member of the SR protein family, 9G8, can activate the splicing of alpha-tropomyosin exon 2. Using RNA affinity chromatography and cross-linking competition assays, we also demonstrate that the heterogeneous nuclear ribonucleoproteins (hnRNPs) H and F bind to and compete for the same elements. Overexpression of hnRNPs H and F blocked 9G8-mediated splicing both in vivo and in vitro, and small interfering RNA-directed depletion of H and F led to an increase in exon 2 splicing. These data suggest that the activation of exon 2 is dependent on the antagonistic activities of 9G8 and hnRNPs H and F.
Figures

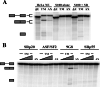
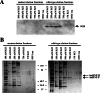
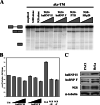
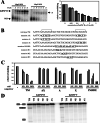
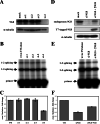
 ). (B) RNA sequences used for cross-linking (TM, AX, A, X, and INSIDE; panels A and C) and in vitro splicing (M1, M2, and M3; panel D) assays. The wild-type sequence consists of the two central exon 2 enhancers (bracketed above) and the sequence between them (shown in Fig. 1A). Nucleotides underlined and in boldface denote mutated nucleotides. (C) UV cross-linking competition. Radiolabeled wild-type TM RNAs (25 nM) were incubated with 9G8 (▪), hnRNP H (□), or hnRNP F (
). (B) RNA sequences used for cross-linking (TM, AX, A, X, and INSIDE; panels A and C) and in vitro splicing (M1, M2, and M3; panel D) assays. The wild-type sequence consists of the two central exon 2 enhancers (bracketed above) and the sequence between them (shown in Fig. 1A). Nucleotides underlined and in boldface denote mutated nucleotides. (C) UV cross-linking competition. Radiolabeled wild-type TM RNAs (25 nM) were incubated with 9G8 (▪), hnRNP H (□), or hnRNP F ( ) in the presence of increasing amounts of cold competitor RNAs. Samples were subjected to UV cross-linking and analyzed as described above. Competitor RNAs were incubated at the indicated molar excess to radiolabeled RNA. RNAs are as indicated in panel B (TM, AX, A, X, and INSIDE). At least three independent competitions were performed, and averages and standard errors are as shown. (D) In vitro splicing reactions were performed with the enhancer-containing dsx-TM and the dsx-M1, M2, M3, and AX mutants in the presence of 9G8 alone (1.5 μM) or when combined with hnRNPs H or F (3 μM).
) in the presence of increasing amounts of cold competitor RNAs. Samples were subjected to UV cross-linking and analyzed as described above. Competitor RNAs were incubated at the indicated molar excess to radiolabeled RNA. RNAs are as indicated in panel B (TM, AX, A, X, and INSIDE). At least three independent competitions were performed, and averages and standard errors are as shown. (D) In vitro splicing reactions were performed with the enhancer-containing dsx-TM and the dsx-M1, M2, M3, and AX mutants in the presence of 9G8 alone (1.5 μM) or when combined with hnRNPs H or F (3 μM).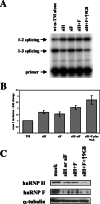
Similar articles
-
hnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of beta-tropomyosin exon 6B.J Biol Chem. 2004 Sep 10;279(37):38249-59. doi: 10.1074/jbc.M405377200. Epub 2004 Jun 18. J Biol Chem. 2004. PMID: 15208309
-
The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20.J Biol Chem. 2003 Aug 29;278(35):32943-53. doi: 10.1074/jbc.M301090200. Epub 2003 Jun 24. J Biol Chem. 2003. PMID: 12826680
-
The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A.EMBO J. 1997 Apr 1;16(7):1772-84. doi: 10.1093/emboj/16.7.1772. EMBO J. 1997. PMID: 9130721 Free PMC article.
-
Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon.RNA. 1998 Jan;4(1):85-100. RNA. 1998. PMID: 9436911 Free PMC article.
-
Tissue-specific splicing of two mutually exclusive exons of the chicken beta-tropomyosin pre-mRNA: positive and negative regulations.Biochimie. 1996;78(6):457-65. doi: 10.1016/0300-9084(96)84752-3. Biochimie. 1996. PMID: 8915535 Review.
Cited by
-
hnRNP A1 and hnRNP H can collaborate to modulate 5' splice site selection.RNA. 2010 Jan;16(1):228-38. doi: 10.1261/rna.1890310. Epub 2009 Nov 19. RNA. 2010. PMID: 19926721 Free PMC article.
-
Pre-mRNA mis-splicing of sarcomeric genes in heart failure.Biochim Biophys Acta Mol Basis Dis. 2017 Aug;1863(8):2056-2063. doi: 10.1016/j.bbadis.2016.11.008. Epub 2016 Nov 5. Biochim Biophys Acta Mol Basis Dis. 2017. PMID: 27825848 Free PMC article. Review.
-
Relative strength of 5' splice-site strength defines functions of SRSF2 and SRSF6 in alternative splicing of Bcl-x pre-mRNA.BMB Rep. 2021 Mar;54(3):176-181. doi: 10.5483/BMBRep.2021.54.3.170. BMB Rep. 2021. PMID: 33050987 Free PMC article.
-
hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc.Mol Cell Biol. 2008 Sep;28(17):5403-19. doi: 10.1128/MCB.00739-08. Epub 2008 Jun 23. Mol Cell Biol. 2008. PMID: 18573884 Free PMC article.
-
HNRNPH1-dependent splicing of a fusion oncogene reveals a targetable RNA G-quadruplex interaction.RNA. 2019 Dec;25(12):1731-1750. doi: 10.1261/rna.072454.119. Epub 2019 Sep 11. RNA. 2019. PMID: 31511320 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
