The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation
- PMID: 16959964
- PMCID: PMC1594588
- DOI: 10.1128/MMBR.00011-06
The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation
Abstract
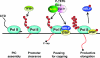
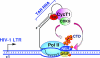
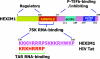
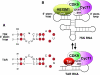
The positive transcription elongation factor b (P-TEFb) stimulates transcriptional elongation by phosphorylating the carboxy-terminal domain of RNA polymerase II and antagonizing the effects of negative elongation factors. Not only is P-TEFb essential for transcription of the vast majority of cellular genes, but it is also a critical host cellular cofactor for the expression of the human immunodeficiency virus (HIV) type 1 genome. Given its important role in globally affecting transcription, P-TEFb's activity is dynamically controlled by both positive and negative regulators in order to achieve a functional equilibrium in sync with the overall transcriptional demand as well as the proliferative state of cells. Notably, this equilibrium can be shifted toward either the active or inactive state in response to diverse physiological stimuli that can ultimately affect the cellular decision between growth and differentiation. In this review, we examine the mechanisms by which the recently identified positive (the bromodomain protein Brd4) and negative (the noncoding 7SK small nuclear RNA and the HEXIM1 protein) regulators of P-TEFb affect the P-TEFb-dependent transcriptional elongation. We also discuss the consequences of perturbations of the dynamic associations of these regulators with P-TEFb in relation to the pathogenesis and progression of several major human diseases, such as cardiac hypertrophy, breast cancer, and HIV infection.
Figures

Similar articles
-
Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation.Mol Cell Biol. 2006 Oct;26(19):7068-76. doi: 10.1128/MCB.00778-06. Mol Cell Biol. 2006. PMID: 16980611 Free PMC article.
-
Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4.Mol Cell. 2005 Aug 19;19(4):535-45. doi: 10.1016/j.molcel.2005.06.029. Mol Cell. 2005. PMID: 16109377
-
Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein.J Biol Chem. 2012 Oct 19;287(43):36609-16. doi: 10.1074/jbc.M112.410746. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952229 Free PMC article.
-
[Misregulation of P-TEFb activity: pathological consequences].Med Sci (Paris). 2012 Feb;28(2):200-5. doi: 10.1051/medsci/2012282019. Epub 2012 Feb 27. Med Sci (Paris). 2012. PMID: 22377309 Review. French.
-
Brd4 and HEXIM1: multiple roles in P-TEFb regulation and cancer.Biomed Res Int. 2014;2014:232870. doi: 10.1155/2014/232870. Epub 2014 Jan 29. Biomed Res Int. 2014. PMID: 24592384 Free PMC article. Review.
Cited by
-
BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells.Oncotarget. 2015 Jul 10;6(19):17698-712. doi: 10.18632/oncotarget.4131. Oncotarget. 2015. PMID: 25989842 Free PMC article.
-
Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism.Elife. 2015 Jun 17;4:e06535. doi: 10.7554/eLife.06535. Elife. 2015. PMID: 26083714 Free PMC article.
-
Confronting proviral HIV infection.Curr HIV/AIDS Rep. 2007 May;4(2):60-4. doi: 10.1007/s11904-007-0009-6. Curr HIV/AIDS Rep. 2007. PMID: 17547826 Review.
-
Association of Tat with promoters of PTEN and PP2A subunits is key to transcriptional activation of apoptotic pathways in HIV-infected CD4+ T cells.PLoS Pathog. 2010 Sep 16;6(9):e1001103. doi: 10.1371/journal.ppat.1001103. PLoS Pathog. 2010. PMID: 20862322 Free PMC article.
-
Down-regulation of cardiac lineage protein (CLP-1) expression in CLP-1 +/- mice affords.J Cell Mol Med. 2009 Aug;13(8B):2744-2753. doi: 10.1111/j.1582-4934.2008.00404.x. J Cell Mol Med. 2009. PMID: 18624753 Free PMC article.
References
-
- Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. - PubMed
-
- Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. - PubMed
-
- Bellan, C., G. De Falco, S. Lazzi, P. Micheli, S. Vicidomini, K. Schurfeld, T. Amato, A. Palumbo, L. Bagella, E. Sabattini, S. Bartolommei, M. Hummel, S. Pileri, P. Tosi, L. Leoncini, and A. Giordano. 2004. CDK9/CYCLIN T1 expression during normal lymphoid differentiation and malignant transformation. J. Pathol. 203:946-952. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

