The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference
- PMID: 16948865
- PMCID: PMC1569866
- DOI: 10.1186/1742-4690-3-57
The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference
Abstract
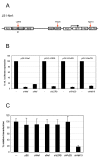
Background: RNA interference (RNAi) has proven to be a powerful tool to suppress gene expression and can be used as a therapeutic strategy against human pathogenic viruses such as human immunodeficiency virus type 1 (HIV-1). Theoretically, RNAi-mediated inhibition can occur at two points in the replication cycle, upon viral entry before reverse transcription of the RNA genome, and on the newly transcribed viral RNA transcripts. There have been conflicting results on whether RNAi can target the RNA genome of infecting HIV-1 particles. We have addressed this issue with HIV-1-based lentiviral vectors.
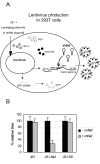
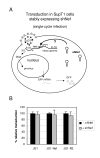
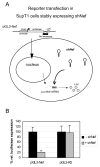
Results: We determined the transduction efficiency of a lentiviral vector, as measured by GFP expressing cells, which reflects the number of successful integration events in a cell line stably expressing shNef. We did not observe a difference in the transduction efficiency comparing lentiviral vectors with or without the Nef target sequence in their genome. The results were similar with particles pseudotyped with either the VSV-G or HIV-1 envelope. Additionally, no reduced transduction efficiencies were observed with multiple other shRNAs targeting the vector genome or with synthetic siNef when transiently transfected prior to transduction.
Conclusion: Our findings indicate that the incoming HIV-1 RNA genome is not targeted by RNAi, probably due to inaccessibility to the RNAi machinery. Thus, therapeutic RNAi strategies aimed at preventing proviral integration should be targeting cellular receptors or co-factors involved in pre-integration events.
Figures

Similar articles
-
Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition.J Virol. 2004 Mar;78(5):2601-5. doi: 10.1128/jvi.78.5.2601-2605.2004. J Virol. 2004. PMID: 14963165 Free PMC article.
-
Silencing of HIV-1 with RNA interference: a multiple shRNA approach.Mol Ther. 2006 Dec;14(6):883-92. doi: 10.1016/j.ymthe.2006.07.007. Epub 2006 Sep 7. Mol Ther. 2006. PMID: 16959541
-
Lentiviral siRNAs targeting multiple highly conserved RNA sequences of human immunodeficiency virus type 1.Gene Ther. 2005 Jul;12(14):1133-44. doi: 10.1038/sj.gt.3302509. Gene Ther. 2005. PMID: 15750613
-
Anti-HIV-1 gene expressing lentiviral vectors as an adjunctive therapy for HIV-1 infection.Curr HIV Res. 2004 Apr;2(2):185-91. doi: 10.2174/1570162043484906. Curr HIV Res. 2004. PMID: 15078182 Review.
-
Lentivirus-mediated RNA interference therapy for human immunodeficiency virus type 1 infection.Hum Gene Ther. 2006 May;17(5):479-86. doi: 10.1089/hum.2006.17.479. Hum Gene Ther. 2006. PMID: 16716105 Review.
Cited by
-
Inhibition of HIV-1 replication using the CRISPR/cas9-no NLS system as a prophylactic strategy.Heliyon. 2022 Aug 31;8(9):e10483. doi: 10.1016/j.heliyon.2022.e10483. eCollection 2022 Sep. Heliyon. 2022. PMID: 36158108 Free PMC article.
-
Retrovirus infected cells contain viral microRNAs.Retrovirology. 2013 Feb 7;10:15. doi: 10.1186/1742-4690-10-15. Retrovirology. 2013. PMID: 23391025 Free PMC article.
-
Inhibition of HIV-1 infection by a unique short hairpin RNA to chemokine receptor 5 delivered into macrophages through hematopoietic progenitor cell transduction.J Gene Med. 2010 Mar;12(3):255-65. doi: 10.1002/jgm.1440. J Gene Med. 2010. PMID: 20186995 Free PMC article.
-
In silico modeling indicates the development of HIV-1 resistance to multiple shRNA gene therapy differs to standard antiretroviral therapy.Retrovirology. 2010 Oct 9;7:83. doi: 10.1186/1742-4690-7-83. Retrovirology. 2010. PMID: 20932334 Free PMC article.
-
Targets of small interfering RNA restriction during human immunodeficiency virus type 1 replication.J Virol. 2008 Mar;82(6):2938-51. doi: 10.1128/JVI.02126-07. Epub 2008 Jan 16. J Virol. 2008. PMID: 18199654 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

